Transcriptional Control of the Lateral-Flagellar Genes of Bradyrhizobium diazoefficiens
- PMID: 28533217
- PMCID: PMC5512216
- DOI: 10.1128/JB.00253-17
Transcriptional Control of the Lateral-Flagellar Genes of Bradyrhizobium diazoefficiens
Abstract
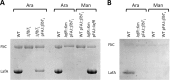
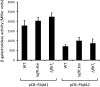
Bradyrhizobium diazoefficiens, a soybean N2-fixing symbiont, possesses a dual flagellar system comprising a constitutive subpolar flagellum and inducible lateral flagella. Here, we analyzed the genomic organization and biosynthetic regulation of the lateral-flagellar genes. We found that these genes are located in a single genomic cluster, organized in two monocistronic transcriptional units and three operons, one possibly containing an internal transcription start site. Among the monocistronic units is blr6846, homologous to the class IB master regulators of flagellum synthesis in Brucella melitensis and Ensifer meliloti and required for the expression of all the lateral-flagellar genes except lafA2, whose locus encodes a single lateral flagellin. We therefore named blr6846 lafR (
Keywords: Bradyrhizobium; FlbT; LafR; expression; flagella; flbT; lafR.
Copyright © 2017 American Society for Microbiology.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

