The Evolutionary Basis of Translational Accuracy in Plants
- PMID: 28533334
- PMCID: PMC5499143
- DOI: 10.1534/g3.117.040626
The Evolutionary Basis of Translational Accuracy in Plants
Abstract
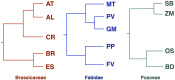
Mistranslation errors compromise fitness by wasting resources on nonfunctional proteins. In order to reduce the cost of mistranslations, natural selection chooses the most accurately translated codons at sites that are particularly important for protein structure and function. We investigated the determinants underlying selection for translational accuracy in several species of plants belonging to three clades: Brassicaceae, Fabidae, and Poaceae. Although signatures of translational selection were found in genes from a wide range of species, the underlying factors varied in nature and intensity. Indeed, the degree of synonymous codon bias at evolutionarily conserved sites varied among plant clades while remaining uniform within each clade. This is unlikely to solely reflect the diversity of tRNA pools because there is little correlation between synonymous codon bias and tRNA abundance, so other factors must affect codon choice and translational accuracy in plant genes. Accordingly, synonymous codon choice at a given site was affected not only by the selection pressure at that site, but also its participation in protein domains or mRNA secondary structures. Although these effects were detected in all the species we analyzed, their impact on translation accuracy was distinct in evolutionarily distant plant clades. The domain effect was found to enhance translational accuracy in dicot and monocot genes with a high GC content, but to oppose the selection of more accurate codons in monocot genes with a low GC content.
Keywords: RNA folding; codon bias; protein domains; translational accuracy.
Copyright © 2017 Camiolo et al.
Figures
References
-
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
