Structure-based prediction of Wnt binding affinities for Frizzled-type cysteine-rich domains
- PMID: 28533339
- PMCID: PMC5500790
- DOI: 10.1074/jbc.M117.786269
Structure-based prediction of Wnt binding affinities for Frizzled-type cysteine-rich domains
Abstract
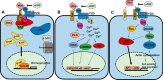
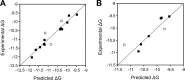
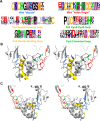
Wnt signaling pathways are of significant interest in development and oncogenesis. The first step in these pathways typically involves the binding of a Wnt protein to the cysteine-rich domain (CRD) of a Frizzled receptor. Wnt-Frizzled interactions can be antagonized by secreted Frizzled-related proteins (SFRPs), which also contain a Frizzled-like CRD. The large number of Wnts, Frizzleds, and SFRPs, as well as the hydrophobic nature of Wnt, poses challenges to laboratory-based investigations of interactions involving Wnt. Here, utilizing structural knowledge of a representative Wnt-Frizzled CRD interaction, as well as experimentally determined binding affinities for a selection of Wnt-Frizzled CRD interactions, we generated homology models of Wnt-Frizzled CRD interactions and developed a quantitative structure-activity relationship for predicting their binding affinities. The derived model incorporates a small selection of terms derived from scoring functions used in protein-protein docking, as well as an energetic term considering the contribution made by the lipid of Wnt to the Wnt-Frizzled binding affinity. Validation with an external test set suggests that the model can accurately predict binding affinity for 75% of cases and that the error associated with the predictions is comparable with the experimental error. The model was applied to predict the binding affinities of the full range of mouse and human Wnt-Frizzled and Wnt-SFRP interactions, indicating trends in Wnt binding affinity for Frizzled and SFRP CRDs. The comprehensive predictions made in this study provide the basis for laboratory-based studies of previously unexplored Wnt-Frizzled and Wnt-SFRP interactions, which, in turn, may reveal further Wnt signaling pathways.
Keywords: Frizzled receptor; Wnt signaling; cysteine-rich domain; homology modeling; lipid-protein interaction; protein-protein interaction; structural biology; structure-activity relationship.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

