Visualization and characterization of individual type III protein secretion machines in live bacteria
- PMID: 28533372
- PMCID: PMC5468683
- DOI: 10.1073/pnas.1705823114
Visualization and characterization of individual type III protein secretion machines in live bacteria
Abstract
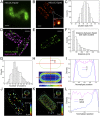
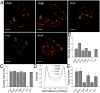
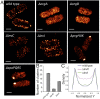
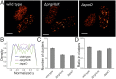
Type III protein secretion machines have evolved to deliver bacterially encoded effector proteins into eukaryotic cells. Although electron microscopy has provided a detailed view of these machines in isolation or fixed samples, little is known about their organization in live bacteria. Here we report the visualization and characterization of the Salmonella type III secretion machine in live bacteria by 2D and 3D single-molecule switching superresolution microscopy. This approach provided access to transient components of this machine, which previously could not be analyzed. We determined the subcellular distribution of individual machines, the stoichiometry of the different components of this machine in situ, and the spatial distribution of the substrates of this machine before secretion. Furthermore, by visualizing this machine in Salmonella mutants we obtained major insights into the machine's assembly. This study bridges a major resolution gap in the visualization of this nanomachine and may serve as a paradigm for the examination of other bacterially encoded molecular machines.
Keywords: Salmonella virulence; bacterial nanomachines; bacterial pathogenesis; protein secretion; superresolution microscopy.
Conflict of interest statement
Conflict of interest statement: J.B. discloses significant financial interest in Bruker Corp. and Hamamatsu Photonics.
Figures

References
-
- Cornelis GR. The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biol Chem. 2010;391:745–751. - PubMed
-
- Charro N, Mota LJ. Approaches targeting the type III secretion system to treat or prevent bacterial infections. Expert Opin Drug Discov. 2015;10:373–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

