Metabarcoding monitoring analysis: the pros and cons of using co-extracted environmental DNA and RNA data to assess offshore oil production impacts on benthic communities
- PMID: 28533985
- PMCID: PMC5437860
- DOI: 10.7717/peerj.3347
Metabarcoding monitoring analysis: the pros and cons of using co-extracted environmental DNA and RNA data to assess offshore oil production impacts on benthic communities
Abstract
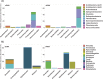
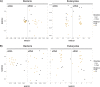
Sequencing environmental DNA (eDNA) is increasingly being used as an alternative to traditional morphological-based identification to characterize biological assemblages and monitor anthropogenic impacts in marine environments. Most studies only assess eDNA which, compared to eRNA, can persist longer in the environment after cell death. Therefore, eRNA may provide a more immediate census of the environment due to its relatively weaker stability, leading some researchers to advocate for the use of eRNA as an additional, or perhaps superior proxy for portraying ecological changes. A variety of pre-treatment techniques for screening eDNA and eRNA derived operational taxonomic units (OTUs) have been employed prior to statistical analyses, including removing singleton taxa (i.e., OTUs found only once) and discarding those not present in both eDNA and eRNA datasets. In this study, we used bacterial (16S ribosomal RNA gene) and eukaryotic (18S ribosomal RNA gene) eDNA- and eRNA-derived data from benthic communities collected at increasing distances along a transect from an oil production platform (Taranaki, New Zealand). Macro-infauna (visual classification of benthic invertebrates) and physico-chemical data were analyzed in parallel. We tested the effect of removing singleton taxa, and removing taxa not present in the eDNA and eRNA libraries from the same environmental sample (trimmed by shared OTUs), by comparing the impact of the oil production platform on alpha- and beta-diversity of the eDNA/eRNA-based biological assemblages, and by correlating these to the morphologically identified macro-faunal communities and the physico-chemical data. When trimmed by singletons, presence/absence information from eRNA data represented the best proxy to detect changes on species diversity for both bacteria and eukaryotes. However, assessment of quantitative beta-diversity from read abundance information of bacteria eRNA did not, contrary to eDNA, reveal any impact from the oil production activity. Overall, the data appeared more robust when trimmed by shared OTUs, showing a greater effect of the platform on alpha- and beta-diversity. Trimming by shared OTUs likely removes taxa derived from legacy DNA and technical artefacts introduced through reverse transcriptase, polymerase-chain-reaction and sequencing. Findings from our scoping study suggest that metabarcoding-based biomonitoring surveys should, if funds, time and expertise allow, be assessed using both eDNA and eRNA products.
Keywords: Bacteria (16S); Benthic ecology; Biomonitoring; Eukaryotes (18S); High-throughput sequencing; Method testing; Oil and gas activities; eDNA; eRNA.
Conflict of interest statement
Dr. Xavier Pochon is an Academic Editor for PeerJ.
Figures


References
-
- Abad D, Albaina A, Aguirre M, Laza-Martínez A, Uriarte I, Iriarte A, Villate F, Estonba A. Is metabarcoding suitable for estuarine plankton monitoring? A comparative study with microscopy. Marine Biology. 2016;163 doi: 10.1007/s00227-016-2920-0. Article 149. - DOI
-
- Bálint M, Bahram M, Eren AM, Faust K, Fuhrman JA, Lindahl B, O’Hara RB, Öpik M, Sogin ML, Unterseher M, Tedersoo L. Millions of reads, thousands of taxa: microbial community structure and associations analyzed via marker genes. FEMS Microbiology Reviews. 2016;40(5):686–700. doi: 10.1093/femsre/fuw017. - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

