Clinical exome sequencing reports: current informatics practice and future opportunities
- PMID: 28535206
- PMCID: PMC7651976
- DOI: 10.1093/jamia/ocx048
Clinical exome sequencing reports: current informatics practice and future opportunities
Abstract
The increased adoption of clinical whole exome sequencing (WES) has improved the diagnostic yield for patients with complex genetic conditions. However, the informatics practice for handling information contained in whole exome reports is still in its infancy, as evidenced by the lack of a common vocabulary within clinical sequencing reports generated across genetic laboratories. Genetic testing results are mostly transmitted using portable document format, which can make secondary analysis and data extraction challenging. This paper reviews a sample of clinical exome reports generated by Clinical Laboratory Improvement Amendments-certified genetic testing laboratories at tertiary-care facilities to assess and identify common data elements. Like structured radiology reports, which enable faster information retrieval and reuse, structuring genetic information within clinical WES reports would help facilitate integration of genetic information into electronic health records and enable retrospective research on the clinical utility of WES. We identify elements listed as mandatory according to practice guidelines but are currently missing from some of the clinical reports, which might help to organize the data when stored within structured databases. We also highlight elements, such as patient consent, that, although they do not appear within any of the current reports, may help in interpreting some of the information within the reports. Integrating genetic and clinical information would assist the adoption of personalized medicine for improved patient care and outcomes.
Keywords: Common data elements; Health Level-7 Fast Healthcare Interoperability Resources; clinical WES; electronic health records; exome report; structured vocabulary.
© The Author 2017. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
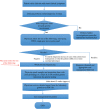

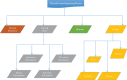
Similar articles
-
Understanding variations in secondary findings reporting practices across U.S. genome sequencing laboratories.AJOB Empir Bioeth. 2018 Jan-Mar;9(1):48-57. doi: 10.1080/23294515.2017.1405095. Epub 2017 Dec 21. AJOB Empir Bioeth. 2018. PMID: 29131714
-
Outcome of Whole Exome Sequencing for Diagnostic Odyssey Cases of an Individualized Medicine Clinic: The Mayo Clinic Experience.Mayo Clin Proc. 2016 Mar;91(3):297-307. doi: 10.1016/j.mayocp.2015.12.018. Mayo Clin Proc. 2016. PMID: 26944241
-
The clinical utility of whole-exome sequencing in the context of rare diseases - the changing tides of medical practice.Clin Genet. 2015 Oct;88(4):313-9. doi: 10.1111/cge.12546. Epub 2015 Jan 6. Clin Genet. 2015. PMID: 25421945 Review.
-
A survey of current practices for genomic sequencing test interpretation and reporting processes in US laboratories.Genet Med. 2017 May;19(5):575-582. doi: 10.1038/gim.2016.152. Epub 2016 Nov 3. Genet Med. 2017. PMID: 27811861 Free PMC article.
-
Regulating whole exome sequencing as a diagnostic test.Hum Genet. 2016 Jun;135(6):655-73. doi: 10.1007/s00439-016-1677-3. Epub 2016 May 11. Hum Genet. 2016. PMID: 27167135 Review.
Cited by
-
FHIR Lab Reports: using SMART on FHIR and CDS Hooks to increase the clinical utility of pharmacogenomic laboratory test results.AMIA Jt Summits Transl Sci Proc. 2020 May 30;2020:683-692. eCollection 2020. AMIA Jt Summits Transl Sci Proc. 2020. PMID: 32477691 Free PMC article.
-
Ethical Considerations on Pediatric Genetic Testing Results in Electronic Health Records.Appl Clin Inform. 2020 Oct;11(5):755-763. doi: 10.1055/s-0040-1718753. Epub 2020 Nov 11. Appl Clin Inform. 2020. PMID: 33176390 Free PMC article.
-
Interoperable genetic lab test reports: mapping key data elements to HL7 FHIR specifications and professional reporting guidelines.J Am Med Inform Assoc. 2021 Nov 25;28(12):2617-2625. doi: 10.1093/jamia/ocab201. J Am Med Inform Assoc. 2021. PMID: 34569596 Free PMC article.
-
Reviewed and updated Algorithm for Genetic Characterization of Syndromic Obesity Phenotypes.Curr Genomics. 2022 Jul 5;23(3):147-162. doi: 10.2174/1389202923666220426093436. Curr Genomics. 2022. PMID: 36777005 Free PMC article. Review.
-
Genomic architecture of fetal central nervous system anomalies using whole-genome sequencing.NPJ Genom Med. 2022 May 13;7(1):31. doi: 10.1038/s41525-022-00301-4. NPJ Genom Med. 2022. PMID: 35562572 Free PMC article.
References
-
- Grada A, Weinbrecht K. Next-generation sequencing: methodology and application. J Invest Dermatol. 2013;133:e11. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

