Parent-offspring regression to estimate the heritability of an HIV-1 trait in a realistic setup
- PMID: 28535768
- PMCID: PMC5442860
- DOI: 10.1186/s12977-017-0356-3
Parent-offspring regression to estimate the heritability of an HIV-1 trait in a realistic setup
Abstract
Background: Parent-offspring (PO) regression is a central tool to determine the heritability of phenotypic traits; i.e., the relative extent to which those traits are controlled by genetic factors. The applicability of PO regression to viral traits is unclear because the direction of viral transmission-who is the donor (parent) and who is the recipient (offspring)-is typically unknown and viral phylogenies are sparsely sampled.
Methods: We assessed the applicability of PO regression in a realistic setting using Ornstein-Uhlenbeck simulated data on phylogenies built from 11,442 Swiss HIV Cohort Study (SHCS) partial pol sequences and set-point viral load (SPVL) data from 3293 patients.
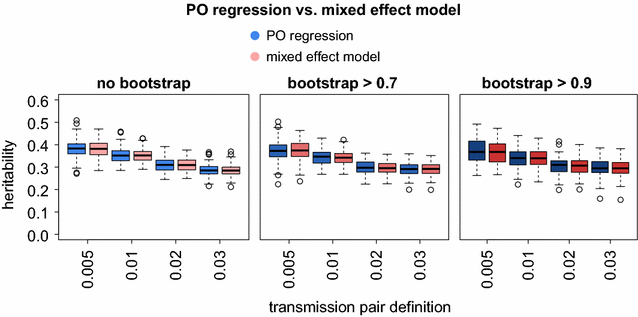
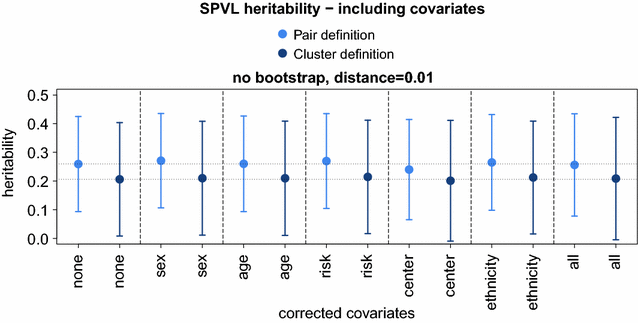
Results: We found that the misidentification of donor and recipient plays a minor role in estimating heritability and showed that sparse sampling does not influence the mean heritability estimated by PO regression. A mixed-effect model approach yielded the same heritability as PO regression but could be extended to clusters of size greater than 2 and allowed for the correction of confounding effects. Finally, we used both methods to estimate SPVL heritability in the SHCS. We employed a wide range of transmission pair criteria to measure heritability and found a strong dependence of the heritability estimates to these criteria. For the most conservative genetic distance criteria, for which heritability estimates are conceptually expected to be closest to true heritability, we found estimates ranging from 32 to 46% across different bootstrap criteria. For less conservative distance criteria, we found estimates ranging down to 8%. All estimates did not change substantially after adjusting for host-demographic factors in the mixed-effect model (±2%).
Conclusions: For conservative transmission pair criteria, both PO regression and mixed-effect models are flexible and robust tools to estimate the contribution of viral genetic effects to viral traits under real-world settings. Overall, we find a strong effect of viral genetics on SPVL that is not confounded by host demographics.
Keywords: HIV-1; Heritability; Mixed-effect model; Ornstein–Uhlenbeck process; Parent-offspring regression; Set-point viral load.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

