N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration
- PMID: 28535802
- PMCID: PMC5442648
- DOI: 10.1186/s13059-017-1224-0
N-BLR, a primate-specific non-coding transcript leads to colorectal cancer invasion and migration
Abstract
Background: Non-coding RNAs have been drawing increasing attention in recent years as functional data suggest that they play important roles in key cellular processes. N-BLR is a primate-specific long non-coding RNA that modulates the epithelial-to-mesenchymal transition, facilitates cell migration, and increases colorectal cancer invasion.
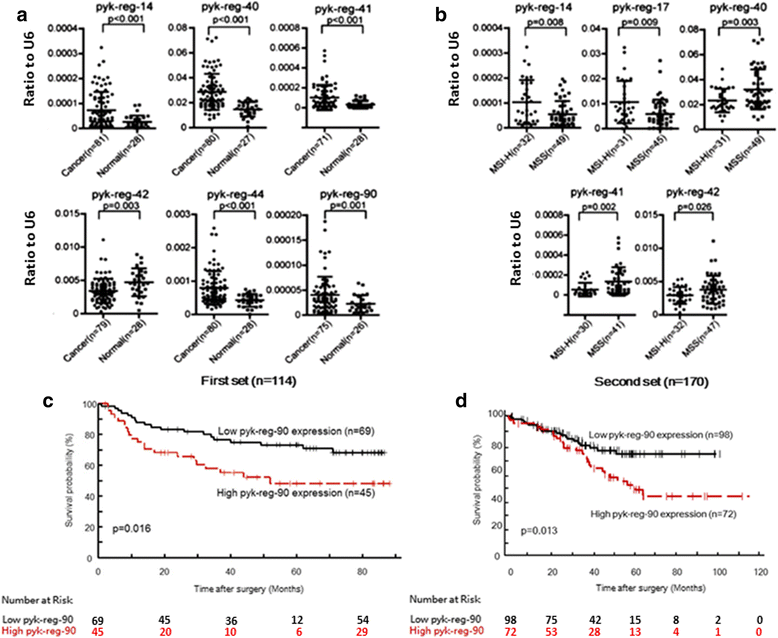
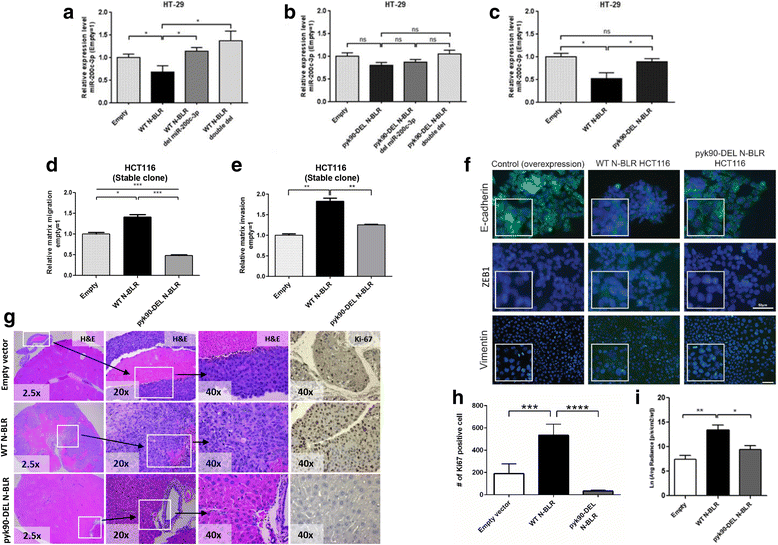
Results: We performed multivariate analyses of data from two independent cohorts of colorectal cancer patients and show that the abundance of N-BLR is associated with tumor stage, invasion potential, and overall patient survival. Through in vitro and in vivo experiments we found that N-BLR facilitates migration primarily via crosstalk with E-cadherin and ZEB1. We showed that this crosstalk is mediated by a pyknon, a short ~20 nucleotide-long DNA motif contained in the N-BLR transcript and is targeted by members of the miR-200 family. In light of these findings, we used a microarray to investigate the expression patterns of other pyknon-containing genomic loci. We found multiple such loci that are differentially transcribed between healthy and diseased tissues in colorectal cancer and chronic lymphocytic leukemia. Moreover, we identified several new loci whose expression correlates with the colorectal cancer patients' overall survival.
Conclusions: The primate-specific N-BLR is a novel molecular contributor to the complex mechanisms that underlie metastasis in colorectal cancer and a potential novel biomarker for this disease. The presence of a functional pyknon within N-BLR and the related finding that many more pyknon-containing genomic loci in the human genome exhibit tissue-specific and disease-specific expression suggests the possibility of an alternative class of biomarkers and therapeutic targets that are primate-specific.
Keywords: CLL; CRC; EMT; N-BLR; Non-coding RNA; Pyknons; Transcription; lncRNA; ncRNA.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

