YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment
- PMID: 28536180
- PMCID: PMC5495066
- DOI: 10.1101/gr.215160.116
YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment
Abstract
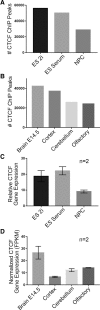
CTCF is an architectural protein with a critical role in connecting higher-order chromatin folding in pluripotent stem cells. Recent reports have suggested that CTCF binding is more dynamic during development than previously appreciated. Here, we set out to understand the extent to which shifts in genome-wide CTCF occupancy contribute to the 3D reconfiguration of fine-scale chromatin folding during early neural lineage commitment. Unexpectedly, we observe a sharp decrease in CTCF occupancy during the transition from naïve/primed pluripotency to multipotent primary neural progenitor cells (NPCs). Many pluripotency gene-enhancer interactions are anchored by CTCF, and its occupancy is lost in parallel with loop decommissioning during differentiation. Conversely, CTCF binding sites in NPCs are largely preexisting in pluripotent stem cells. Only a small number of CTCF sites arise de novo in NPCs. We identify another zinc finger protein, Yin Yang 1 (YY1), at the base of looping interactions between NPC-specific genes and enhancers. Putative NPC-specific enhancers exhibit strong YY1 signal when engaged in 3D contacts and negligible YY1 signal when not in loops. Moreover, siRNA knockdown of Yy1 specifically disrupts interactions between key NPC enhancers and their target genes. YY1-mediated interactions between NPC regulatory elements are often nested within constitutive loops anchored by CTCF. Together, our results support a model in which YY1 acts as an architectural protein to connect developmentally regulated looping interactions; the location of YY1-mediated interactions may be demarcated in development by a preexisting topological framework created by constitutive CTCF-mediated interactions.
© 2017 Beagan et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
