SIRT3-Mediated Dimerization of IDH2 Directs Cancer Cell Metabolism and Tumor Growth
- PMID: 28536275
- PMCID: PMC5540757
- DOI: 10.1158/0008-5472.CAN-16-2393
SIRT3-Mediated Dimerization of IDH2 Directs Cancer Cell Metabolism and Tumor Growth
Abstract
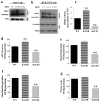
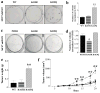
The isocitrate dehydrogenase IDH2 produces α-ketoglutarate by oxidizing isocitrate, linking glucose metabolism to oxidative phosphorylation. In this study, we report that loss of SIRT3 increases acetylation of IDH2 at lysine 413 (IDH2-K413-Ac), thereby decreasing its enzymatic activity by reducing IDH2 dimer formation. Expressing a genetic acetylation mimetic IDH2 mutant (IDH2K413Q) in cancer cells decreased IDH2 dimerization and enzymatic activity and increased cellular reactive oxygen species and glycolysis, suggesting a shift in mitochondrial metabolism. Concurrently, overexpression of IDH2K413Q promoted cell transformation and tumorigenesis in nude mice, resulting in a tumor-permissive phenotype. IHC staining showed that IDH2 acetylation was elevated in high-risk luminal B patients relative to low-risk luminal A patients. Overall, these results suggest a potential relationship between SIRT3 enzymatic activity, IDH2-K413 acetylation-determined dimerization, and a cancer-permissive phenotype. Cancer Res; 77(15); 3990-9. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
The authors have no conflict of interest
Figures

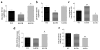

References
-
- Xu X, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279(32):33946–33957. - PubMed
-
- Cupp JR, McAlister-Henn L. Kinetic analysis of NAD(+)-isocitrate dehydrogenase with altered isocitrate binding sites: contribution of IDH1 and IDH2 subunits to regulation and catalysis. Biochemistry. 1993;32(36):9323–9328. - PubMed
-
- Haselbeck RJ, McAlister-Henn L. Function and expression of yeast mitochondrial NAD- and NADP-specific isocitrate dehydrogenases. J Biol Chem. 1993;268(16):12116–12122. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

