The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells
- PMID: 28536419
- PMCID: PMC5442156
- DOI: 10.1038/s41598-017-02357-0
The long non-coding RNA LINC00152 is essential for cell cycle progression through mitosis in HeLa cells
Abstract
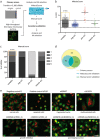
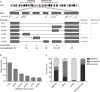
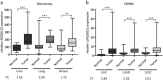
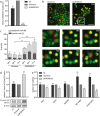
In recent years, long non-coding RNA (lncRNA) research has identified essential roles of these transcripts in virtually all physiological cellular processes including tumorigenesis, but their functions and molecular mechanisms are poorly understood. In this study, we performed a high-throughput siRNA screen targeting 638 lncRNAs deregulated in cancer entities to analyse their impact on cell division by using time-lapse microscopy. We identified 26 lncRNAs affecting cell morphology and cell cycle including LINC00152. This transcript was ubiquitously expressed in many human cell lines and its RNA levels were significantly upregulated in lung, liver and breast cancer tissues. A comprehensive sequence analysis of LINC00152 revealed a highly similar paralog annotated as MIR4435-2HG and several splice variants of both transcripts. The shortest and most abundant isoform preferentially localized to the cytoplasm. Cells depleted of LINC00152 arrested in prometaphase of mitosis and showed reduced cell viability. In RNA affinity purification (RAP) studies, LINC00152 interacted with a network of proteins that were associated with M phase of the cell cycle. In summary, we provide new insights into the properties and biological function of LINC00152 suggesting that this transcript is crucial for cell cycle progression through mitosis and thus, could act as a non-coding oncogene.
Conflict of interest statement
Sven Diederichs is co-owner of the siTOOLs Biotech GmbH. Parts of this study are parts of the PhD theses of L.N. and M.Ga.
Figures
Similar articles
-
Long non-coding RNA LINC00152 is up-regulated in ovarian cancer tissues and regulates proliferation and cell cycle of SKOV3 cells.Eur Rev Med Pharmacol Sci. 2019 Nov;23(22):9803-9813. doi: 10.26355/eurrev_201911_19543. Eur Rev Med Pharmacol Sci. 2019. PMID: 31799647
-
Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer.Oncotarget. 2016 Mar 1;7(9):9773-87. doi: 10.18632/oncotarget.6949. Oncotarget. 2016. PMID: 26799422 Free PMC article.
-
lncRNA LINC00152 knockdown had effects to suppress biological activity of lung cancer via EGFR/PI3K/AKT pathway.Biomed Pharmacother. 2017 Oct;94:644-651. doi: 10.1016/j.biopha.2017.07.120. Epub 2017 Aug 5. Biomed Pharmacother. 2017. PMID: 28787699
-
Improved characterization of the relationship between long intergenic non-coding RNA Linc00152 and the occurrence and development of malignancies.Cancer Med. 2019 Aug;8(10):4722-4731. doi: 10.1002/cam4.2245. Epub 2019 Jul 4. Cancer Med. 2019. PMID: 31270960 Free PMC article. Review.
-
LINC00152: A pivotal oncogenic long non-coding RNA in human cancers.Cell Prolif. 2017 Aug;50(4):e12349. doi: 10.1111/cpr.12349. Epub 2017 May 2. Cell Prolif. 2017. PMID: 28464433 Free PMC article. Review.
Cited by
-
Transcriptome Analysis Identifies LINC00152 as a Biomarker of Early Relapse and Mortality in Acute Lymphoblastic Leukemia.Genes (Basel). 2020 Mar 13;11(3):302. doi: 10.3390/genes11030302. Genes (Basel). 2020. PMID: 32183133 Free PMC article.
-
LncRNA SNHG6 Silencing Could Arrest Progression of High Grade Colorectal Cancers.Iran J Pathol. 2022 Winter;17(1):29-36. doi: 10.30699/IJP.2021.527781.2610. Epub 2021 Dec 15. Iran J Pathol. 2022. PMID: 35096086 Free PMC article.
-
LINC01021 Attenuates Expression and Affects Alternative Splicing of a Subset of p53-Regulated Genes.Cancers (Basel). 2024 Apr 24;16(9):1639. doi: 10.3390/cancers16091639. Cancers (Basel). 2024. PMID: 38730591 Free PMC article.
-
Single-cell transcriptomics identifies multiple pathways underlying antitumor function of TCR- and CD8αβ-engineered human CD4+ T cells.Sci Adv. 2020 Jul 3;6(27):eaaz7809. doi: 10.1126/sciadv.aaz7809. eCollection 2020 Jul. Sci Adv. 2020. PMID: 32923584 Free PMC article.
-
Targeting and engineering long non-coding RNAs for cancer therapy.Nat Rev Genet. 2024 Aug;25(8):578-595. doi: 10.1038/s41576-024-00693-2. Epub 2024 Feb 29. Nat Rev Genet. 2024. PMID: 38424237 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

