Novel highly divergent sapoviruses detected by metagenomics analysis in straw-colored fruit bats in Cameroon
- PMID: 28536431
- PMCID: PMC5520483
- DOI: 10.1038/emi.2017.20
Novel highly divergent sapoviruses detected by metagenomics analysis in straw-colored fruit bats in Cameroon
Abstract
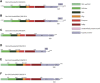
Sapoviruses (SaVs) belong to the Sapovirus genus, in the family Caliciviridae. They have been associated with gastroenteritis in humans and in pigs but not in other animals. In addition, some strains from pigs, chimpanzees and rodents show close sequence identity with human SaVs thereby suggesting the possibility of interspecies transmissions. Bats are known to be a major reservoir of zoonotic viruses, however, very little is known about the genetic diversity of SaVs in bats. To explore the genetic diversity of bat SaVs, fecal samples of Eidolon helvum and Epomophorus gambianus were treated according to the NetoVIR protocol and sequenced by Illumina technology. Nearly complete genome sequences of six highly divergent SaVs and one partial SaV (only VP1 region) were identified in Eidolon helvum and based on sequence identities and phylogenetic analysis, they potentially represent two novel genogroups, only distantly related to known SaVs. Furthermore, comparing these sequences with currently used screening primers and probes indicated that the novel SaVs would not be detected in routine epidemiological screening studies in humans in case an interspecies transmission would occur. Therefore, we designed and validated new primers that can detect both human and bat SaVs. In this study, we identified multiple novel bat SaVs, however, further epidemiological studies in humans are needed to unravel their potential role in gastroenteritis.
Figures

References
-
- Madeley CR, Cosgrove BP. Letter: Caliciviruses in man. Lancet 1976; 1: 199–200. - PubMed
-
- Mayo MA. A summary of taxonomic changes recently approved by ICTV. Arch Virol 2002; 147: 1655–1663. - PubMed
-
- Madeley CR. Comparison of the features of astroviruses and caliciviruses seen in samples of feces by electron microscopy. J Infect Dis 1979; 139: 519–523. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
