NMD monitors translational fidelity 24/7
- PMID: 28536849
- PMCID: PMC5668330
- DOI: 10.1007/s00294-017-0709-4
NMD monitors translational fidelity 24/7
Abstract
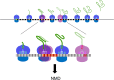
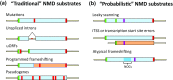
Nonsense-mediated mRNA decay (NMD) is generally thought to be a eukaryotic mRNA surveillance pathway tasked with the elimination of transcripts harboring an in-frame premature termination codon (PTC). As presently conceived, NMD acting in this manner minimizes the likelihood that potentially toxic polypeptide fragments would accumulate in the cytoplasm. This notion is to be contrasted to the results of systematic RNA-Seq and microarray analyses of NMD substrates in multiple model systems, two different experimental approaches which have shown that many mRNAs identified as NMD substrates fail to contain a PTC. Our recent results provide insight into, as well as a possible solution for, this conundrum. By high-resolution profiling of mRNAs that accumulate in yeast when the principal NMD regulatory genes (UPF1, UPF2, and UPF3) are deleted, we identified approximately 900 NMD substrates, the majority of which are normal-looking mRNAs that lack PTCs. Analyses of ribosomal profiling data revealed that the latter mRNAs tended to manifest elevated rates of out-of-frame translation, a phenomenon that would lead to premature translation termination in alternative reading frames. These results, and related observations of heterogeneity in mRNA isoforms, suggest that NMD should be reconsidered as a probabilistic mRNA quality control pathway that is continually active throughout an mRNA's life cycle.
Keywords: Frameshifting; NMD substrates; Probabilistic mRNA decay; Translational fidelity.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

