Transmembrane helix hydrophobicity is an energetic barrier during the retrotranslocation of integral membrane ERAD substrates
- PMID: 28539401
- PMCID: PMC5509421
- DOI: 10.1091/mbc.E17-03-0184
Transmembrane helix hydrophobicity is an energetic barrier during the retrotranslocation of integral membrane ERAD substrates
Abstract
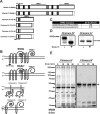
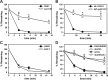
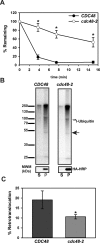
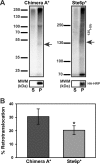
Integral membrane proteins fold inefficiently and are susceptible to turnover via the endoplasmic reticulum-associated degradation (ERAD) pathway. During ERAD, misfolded proteins are recognized by molecular chaperones, polyubiquitinated, and retrotranslocated to the cytoplasm for proteasomal degradation. Although many aspects of this pathway are defined, how transmembrane helices (TMHs) are removed from the membrane and into the cytoplasm before degradation is poorly understood. In this study, we asked whether the hydrophobic character of a TMH acts as an energetic barrier to retrotranslocation. To this end, we designed a dual-pass model ERAD substrate, Chimera A*, which contains the cytoplasmic misfolded domain from a characterized ERAD substrate, Sterile 6* (Ste6p*). We found that the degradation requirements for Chimera A* and Ste6p* are similar, but Chimera A* was retrotranslocated more efficiently than Ste6p* in an in vitro assay in which retrotranslocation can be quantified. We then constructed a series of Chimera A* variants containing synthetic TMHs with a range of ΔG values for membrane insertion. TMH hydrophobicity correlated inversely with retrotranslocation efficiency, and in all cases, retrotranslocation remained Cdc48p dependent. These findings provide insight into the energetic restrictions on the retrotranslocation reaction, as well as a new computational approach to predict retrotranslocation efficiency.
© 2017 Guerriero et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures




References
-
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997.
-
- Amberg DC, Burke D, Strathern JN, Cold Spring Harbor Laboratory . Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005.
-
- Baeza-Delgado C, Marti-Renom MA, Mingarro I. Structure-based statistical analysis of transmembrane helices. Eur Biophys J. 2013;42:199–207. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

