γ-Secretase inhibitors in cancer clinical trials are pharmacologically and functionally distinct
- PMID: 28539479
- PMCID: PMC5494507
- DOI: 10.15252/emmm.201607265
γ-Secretase inhibitors in cancer clinical trials are pharmacologically and functionally distinct
Abstract
γ-Secretase inhibitors (GSIs) are being actively repurposed as cancer therapeutics based on the premise that inhibition of NOTCH1 signaling in select cancers is therapeutic. Using novel assays to probe effects of GSIs against a broader panel of substrates, we demonstrate that clinical GSIs are pharmacologically distinct. GSIs show differential profiles of inhibition of the various NOTCH substrates, with some enhancing cleavage of other NOTCH substrates at concentrations where NOTCH1 cleavage is inhibited. Several GSIs are also potent inhibitors of select signal peptide peptidase (SPP/SPPL) family members. Extending these findings to mammosphere inhibition assays in triple-negative breast cancer lines, we establish that these GSIs have different functional effects. We also demonstrate that the processive γ-secretase cleavage pattern established for amyloid precursor protein (APP) occurs in multiple substrates and that potentiation of γ-secretase cleavage is attributable to a direct action of low concentrations of GSIs on γ-secretase. Such data definitively demonstrate that the clinical GSIs are not biological equivalents, and provide an important framework to evaluate results from ongoing and completed human trials with these compounds.
Keywords: NOTCH; cancer therapy; γ‐secretase inhibitors.
© 2017 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
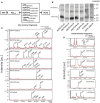
Substrates based on Aβ peptide and γ‐secretase substrate TMDs for cell‐free assay.
Western blot of substrates overexpressed and purified from BL21.
Aβ‐Nβ chimeric peptides (NTF) IP‐MS of recombinant substrates after incubation with CHAPSO‐solubilized CHO cell membrane with (red) or without (black) 1 μM LY411575.
COOH‐terminal fragments (CTFs) IP‐MS of recombinant substrates with (red) or without (black) 1 μM LY411575.


Chemical structure of GSIs used in this work.
5 μM GSIs completely abolished rNOTCH1sub γ‐secretase cleavage.
IP‐MS of NOTCH substrate with (red) or without (black) 25 nM BMS‐906024.
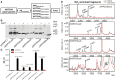
Substrates for cell‐based assay.
Western blot of substrate transfected H4 cell lysate detected with 6E10 antibody. Arrows indicate substrate monomers.
Aβ was detected in culture media using Aβ ELISA, and the secretion was inhibited by GSI. Significance analysis with control (pCDNA3.1) was performed by one‐way ANOVA using GraphPad. Values are mean ± SD of three tests.
Aβ‐like peptides were detected in culture media by IP‐MS (black). 1 μM LY411575 abolished all production (red).
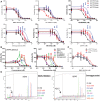
IC50 curves of BMS‐906024, PF‐3084014, RO4929097, Semagacestat, MK‐0752, and DAPT on cNOTCHsub.
Dose responses of cAPPC100sub and full‐length APP cleavage to GSIs. The Aβ level of cells with DMSO was set as 100% activity.
Low BMS‐906024 dose potentiation of cNOTCH1sub, cAPPC100sub, and full‐length APP.
Low‐dose potentiation of cleavage of rAPPC100sub was tested with in vitro assay and IP‐MS. Aβ15 from rAPPC100sub was used as internal standards as shown on the left side of each spectrum.

Confluent MDA‐MB‐231 cells were treated with indicated concentrations of GSIs for 1 h and then with 5 mM EDTA for 5 min to induce NOTCH1 activation. The figure shows Western blot analysis of cell lysates treated with GSIs.
Band intensities of NOTCH1 ICD with 0.1 μM GSIs were normalized for β‐actin using ImageJ Software. Values are mean ± SD of three tests.
APP‐CTF Western blot of MDA‐MB‐231 cells treated with indicated concentration of GSIs for 16 h.
Band intensities of APP‐CTF with 0.1 μM GSIs were normalized for DMSO control group. Values are mean ± SD of three tests.
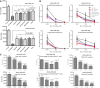
Mammospheres from MDA‐MB‐231 and MDA‐MB‐468 cell lines were treated with 10 μM GSIs for 1 week. Absolute viable cell counts following GSI treatment are shown.
Viable cells (1,000, 100, and 10) from each GSI‐treated conditions were then plated for limiting dilutions assay without further treatments.
Percentage and total number of cancer stem‐like cells (CD44+CD24low) of PF‐3084014‐treated mammospheres were analyzed by flow cytometry.
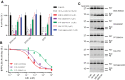
Conditioned media ELISA of HEK 293t cells co‐transfected with FBA and SPPLs. The Aβ level of cells with DMSO was set as 100% activity. Values are mean ± SD of three tests. ***P = 0.003; ****P < 0.0001.
GSIs dose response of FBA/SPPL2b overexpression cells. Values are mean ± SD of three tests.
Turnover of endogenous CD74 P8 is inhibited by RO4929079 and BMS‐906024. Cell lysate Western blot of A20 cells treated with GSIs was developed with In‐1 antibody. MK‐0752 and Semagacestat tests used the same control lane (0 nM).
References
-
- Albright CF, Dockens RC, Meredith JE Jr, Olson RE, Slemmon R, Lentz KA, Wang JS, Denton RR, Pilcher G, Rhyne PW et al (2013) Pharmacodynamics of selective inhibition of gamma‐secretase by avagacestat. J Pharmacol Exp Ther 344: 686–695 - PubMed
-
- Andersson ER, Lendahl U (2014) Therapeutic modulation of Notch signalling–are we there yet? Nat Rev Drug Discov 13: 357–378 - PubMed
-
- Azzam DJ, Zhao D, Sun J, Minn AJ, Ranganathan P, Drews‐Elger K, Han X, Picon‐Ruiz M, Gilbert CA, Wander SA et al (2013) Triple negative breast cancer initiating cell subsets differ in functional and molecular characteristics and in gamma‐secretase inhibitor drug responses. EMBO Mol Med 5: 1502–1522 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

