Phylogenetic analysis of Monascus and new species from honey, pollen and nests of stingless bees
- PMID: 28539687
- PMCID: PMC5432663
- DOI: 10.1016/j.simyco.2017.04.001
Phylogenetic analysis of Monascus and new species from honey, pollen and nests of stingless bees
Abstract
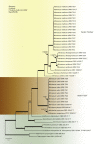
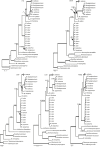
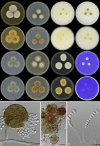
The genus Monascus was described by van Tieghem (1884) to accommodate M. ruber and M. mucoroides, two species with non-ostiolate ascomata. Species delimitation in the genus is still mainly based on phenotypic characters, and taxonomic studies that include sequence data are limited. The genus is of economic importance. Species are used in fermented Asian foods as food colourants (e.g. 'red rice' (ang-kak, angka)) and found as spoilage organisms, and recently Monascus was found to be essential in the lifecycle of stingless bees. In this study, a polyphasic approach was applied combining morphological characters, ITS, LSU, β-tubulin, calmodulin and RNA polymerase II second largest subunit sequences and extrolite data, to delimit species and to study phylogenetic relationships in Monascus. Furthermore, 30 Monascus isolates from honey, pollen and nests of stingless bees in Brazil were included. Based on this polyphasic approach, the genus Monascus is resolved in nine species, including three new species associated with stingless bees (M. flavipigmentosus sp. nov., M. mellicola sp. nov., M. recifensis sp. nov., M. argentinensis, M. floridanus, M. lunisporas, M. pallens, M. purpureus, M. ruber), and split in two new sections (section Floridani sect. nov., section Rubri sect. nov.). Phylogenetic analysis showed that the xerophile Monascus eremophilus does not belong in Monascus and monophyly in Monascus is restored with the transfer of M. eremophilus to Penicillium (P. eremophilum comb. nov.). A list of accepted and excluded Monascus and Basipetospora species is given, together with information on (ex-)types cultures and barcode sequence data.
Keywords: Aspergillaceae; Extrolites; Fungal ecology; Monascus flavipigmentosus R.N. Barbosa, Souza-Motta, N.T. Oliveira & Houbraken; Monascus mellicola R.N. Barbosa, Souza-Motta, N.T. Oliveira & Houbraken; Monascus recifensis R.N. Barbosa, Souza-Motta, N.T. Oliveira & Houbraken; Monascus section Floridani R.N. Barbosa & Houbraken; Monascus section Rubri R.N. Barbosa & Houbraken; Penicillium eremophilum (A.D. Hocking & Pitt) Houbraken, Leong & Vinnere-Pettersson; Phylogeny; Taxonomy.
Figures





References
-
- Arx J.A. von. A re-evaluation of the Eurotiales. Persoonia. 1987;13:273–300.
-
- Barnard E.L., Cannon P.F. A new species of Monascus from pine tissues in Florida. Mycologia. 1987;79:479–484.
-
- Benny G.L., Kimbrough J.W. Synopsis of the orders and families of Plectomycetes with keys to genera. Mycotaxon. 1980;12:1–91.
-
- Berbee M.L., Yoshimura A., Sugiyama J. Is Penicillium monophyletic? An evaluation of phylogeny in the family Trichocomaceae from 18S, 5.8S and ITS ribosomal DNA sequence data. Mycologia. 1995;87:210–222.
-
- Beyma F.H. van. Beschreibung einiger neuer Pilzarten aus dem Centraalbureau voor Schimmelcultures – Baarn (Holland) Zentralblatt für Bakteriologie und Parasitenkunde Abteilung. 1933;88:132–141.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
