Yeast ENV9 encodes a conserved lipid droplet (LD) short-chain dehydrogenase involved in LD morphology
- PMID: 28540421
- PMCID: PMC5802364
- DOI: 10.1007/s00294-017-0702-y
Yeast ENV9 encodes a conserved lipid droplet (LD) short-chain dehydrogenase involved in LD morphology
Abstract
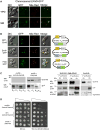
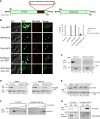
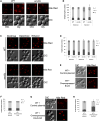
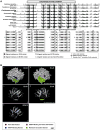
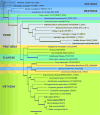
Lipid droplets (LDs) have emerged as dynamic and interactive organelles with important roles in lipid metabolism and membrane biogenesis. Here, we report that Saccharomyces cerevisiae Env9 is a novel conserved oxidoreductase involved in LD morphology. Microscopic and biochemical studies confirm localization of tagged Env9 to LDs and implicate its C-terminal hydrophobic domain (aa241-265) in its membrane association and stability. Confocal studies reveal a role for Env9 in LD morphology. Env9 positively affects both formation of large LDs upon overexpression and LD proliferation under poor carbon source. In silico bioinformatic and modeling approaches establish that ENV9 is a widely conserved member of the short-chain dehydrogenase (SDR) superfamily. Bayesian phylogenetic studies strongly support ENV9 as an ortholog of human SDR retinol dehydrogenase 12 (RDH12). Dehydrogenase activity of Env9 was confirmed by in vitro oxidoreductase assays. RDH12 mutations have been linked to Leber Congenital Amaurosis. Similar site-directed point mutations in the predicted Env9 oxidoreductase active site (N146L) or cofactor-binding site (G23-24A) abolished its reductase activity in vitro, consistent with those reported in other retinol dehydrogenases. The same residues were essential for affecting LD size and number in vivo. Taken together, our results implicate oxidoreductase activity of Env9 in its cellular role in LD morphology.
Keywords: ENV9; LD morphology; LD number; LD size; RDH12 ortholog; Short-chain dehydrogenase.
Figures



References
-
- Beeler T, Bacikova D, Gable K, Hopkins L, Johnson C, Slife H, Dunn T. The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Δ mutant. J Biol Chem. 1998;273:30688–30694. - PubMed
-
- Belyaeva OV, Korkina OV, Stetsenko AV, Kim T, Nelson PS, Kedishvili NY. Biochemical properties of purified human retinol dehydrogenase 12 (RDH12): catalytic efficiency toward retinoids and C9 aldehydes and effects of cellular retinol-binding protein type I (CRBPI) and cellular retinaldehyde-binding protein (CRALBP) on the oxidation and reduction of retinoids. Biochemistry. 2005;44:7035–7047. doi: 10.1021/bi050226k. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

