Finding a helix in a haystack: nucleic acid cytometry with droplet microfluidics
- PMID: 28540956
- PMCID: PMC6005652
- DOI: 10.1039/c7lc00241f
Finding a helix in a haystack: nucleic acid cytometry with droplet microfluidics
Abstract
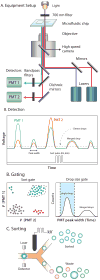
Nucleic acids encode the information of life, programming cellular functions and dictating many biological outcomes. Differentiating between cells based on their nucleic acid programs is, thus, a powerful way to unravel the genetic bases of many phenotypes. This is especially important considering that most cells exist in heterogeneous populations, requiring them to be isolated before they can be studied. Existing flow cytometry techniques, however, are unable to reliably recover specific cells based on nucleic acid content. Nucleic acid cytometry is a new field built on droplet microfluidics that allows robust identification, sorting, and sequencing of cells based on specific nucleic acid biomarkers. This review highlights applications that immediately benefit from the approach, biological questions that can be addressed for the first time with it, and considerations for building successful workflows.
Figures
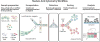

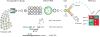

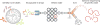
References
-
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RCT, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Nat. Med. 2002;8:68–74. - PubMed
-
- Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, D'Amico M, Pestell RG, West M, Nevins JR. Nat. Genet. 2003;34:226–230. - PubMed
-
- Kintses B, van Vliet LD, Devenish SR, Hollfelder F. Curr. Opin. Chem. Biol. 2010;14:548–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

