Interlinked DNA nano-circles for measuring topoisomerase II activity at the level of single decatenation events
- PMID: 28541438
- PMCID: PMC5570003
- DOI: 10.1093/nar/gkx480
Interlinked DNA nano-circles for measuring topoisomerase II activity at the level of single decatenation events
Abstract
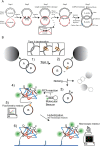
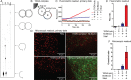
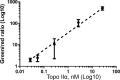
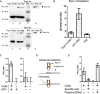
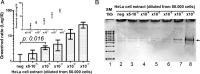
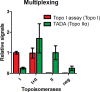
DNA nano-structures present appealing new means for monitoring different molecules. Here, we demonstrate the assembly and utilization of a surface-attached double-stranded DNA catenane composed of two intact interlinked DNA nano-circles for specific and sensitive measurements of the life essential topoisomerase II (Topo II) enzyme activity. Topo II activity was detected via the numeric release of DNA nano-circles, which were visualized at the single-molecule level in a fluorescence microscope upon isothermal amplification and fluorescence labeling. The transition of each enzymatic reaction to a micrometer sized labeled product enabled quantitative detection of Topo II activity at the single decatenation event level rendering activity measurements in extracts from as few as five cells possible. Topo II activity is a suggested predictive marker in cancer therapy and, consequently, the described highly sensitive monitoring of Topo II activity may add considerably to the toolbox of individualized medicine where decisions are based on very sparse samples.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Topoisomerase VI is a chirally-selective, preferential DNA decatenase.Elife. 2022 Jan 25;11:e67021. doi: 10.7554/eLife.67021. Elife. 2022. PMID: 35076393 Free PMC article.
-
The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation.Nucleic Acids Res. 2008 Oct;36(18):5822-31. doi: 10.1093/nar/gkn560. Epub 2008 Sep 12. Nucleic Acids Res. 2008. PMID: 18790802 Free PMC article.
-
Structural determinants of the catalytic inhibition of human topoisomerase IIα by salicylate analogs and salicylate-based drugs.Biochem Pharmacol. 2014 Jun 15;89(4):464-76. doi: 10.1016/j.bcp.2014.03.011. Epub 2014 Mar 30. Biochem Pharmacol. 2014. PMID: 24695359
-
Recent advances in the development of dual topoisomerase I and II inhibitors as anticancer drugs.Curr Med Chem. 2010;17(35):4270-90. doi: 10.2174/092986710793361252. Curr Med Chem. 2010. PMID: 20939813 Review.
-
Simulation of DNA catenanes.Phys Chem Chem Phys. 2009 Dec 7;11(45):10543-52. doi: 10.1039/b910812b. Epub 2009 Oct 23. Phys Chem Chem Phys. 2009. PMID: 20145800 Free PMC article. Review.
Cited by
-
Tricking enzymes in living cells: a mechanism-based strategy for design of DNA topoisomerase biosensors.J Nanobiotechnology. 2021 Dec 7;19(1):407. doi: 10.1186/s12951-021-01155-1. J Nanobiotechnology. 2021. PMID: 34876137 Free PMC article.
-
Constitutively active Artemis nuclease recognizes structures containing single-stranded DNA configurations.DNA Repair (Amst). 2019 Nov;83:102676. doi: 10.1016/j.dnarep.2019.102676. Epub 2019 Jul 26. DNA Repair (Amst). 2019. PMID: 31377101 Free PMC article.
-
Rolling circle RNA synthesis catalyzed by RNA.Elife. 2022 Feb 2;11:e75186. doi: 10.7554/eLife.75186. Elife. 2022. PMID: 35108196 Free PMC article.
-
Advancing Topoisomerase Research Using DNA Nanotechnology.Small Methods. 2025 Jun;9(6):e2401113. doi: 10.1002/smtd.202401113. Epub 2024 Nov 11. Small Methods. 2025. PMID: 39526512 Free PMC article. Review.
References
-
- Halder S., Krishnan Y.. Design of ultrasensitive DNA-based fluorescent pH sensitive nanodevices. Nanoscale. 2015; 7:10008–10012. - PubMed
-
- Brian R Baker, Lai R.Y., Wood M.S., Doctor E.H., Heeger A.J., Plaxco K.W.. An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids. J. Am. Chem. Soc. 2006; 128:3138–3139. - PubMed
-
- Zayats M., Huang Y., Gill R., Ma C.-A., Willner I.. Label-free and reagentless aptamer-based sensors for small molecules. J. Am. Chem. Soc. 2006; 128:13666–13667. - PubMed
-
- Liu J., Wang C., Jiang Y., Hu Y., Li J., Yang S., Li Y., Yang R., Tan W., Huang C.Z.. Graphene signal amplification for sensitive and real-time fluorescence anisotropy detection of small molecules. Anal. Chem. 2013; 85:1424–1430. - PubMed
-
- Bi S., Li L., Zhang S.. Triggered polycatenated DNA scaffolds for DNA sensors and aptasensors by a combination of rolling circle amplification and DNAzyme amplification. Anal. Chem. 2010; 82:9447–9454. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

