Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection
- PMID: 28542207
- PMCID: PMC5441588
- DOI: 10.1371/journal.pone.0177340
Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection
Abstract
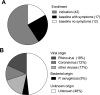
Background: Lung transplant patients are a vulnerable group of immunosuppressed patients that are prone to frequent respiratory infections. We studied 60 episodes of respiratory symptoms in 71 lung transplant patients. Almost half of these episodes were of unknown infectious etiology despite extensive routine diagnostic testing.
Methods: We re-analyzed respiratory samples of all episodes with undetermined etiology in order to detect potential viral pathogens missed/not accounted for in routine diagnostics. Respiratory samples were enriched for viruses by filtration and nuclease digestion, whole nucleic acids extracted and randomly amplified before high throughput metagenomic virus sequencing. Viruses were identified by a bioinformatic pipeline and confirmed and quantified using specific real-time PCR.
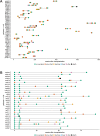
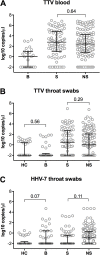
Results: In completion of routine diagnostics, we identified and confirmed a viral etiology of infection by our metagenomic approach in four patients (three Rhinovirus A, one Rhinovirus B infection) despite initial negative results in specific multiplex PCR. Notably, the majority of samples were also positive for Torque teno virus (TTV) and Human Herpesvirus 7 (HHV-7). While TTV viral loads increased with immunosuppression in both throat swabs and blood samples, HHV-7 remained at low levels throughout the observation period and was restricted to the respiratory tract.
Conclusion: This study highlights the potential of metagenomic sequencing for virus diagnostics in cases with previously unknown etiology of infection and in complex diagnostic situations such as in immunocompromised hosts.
Conflict of interest statement
Figures
References
-
- Fisher CE, Preiksaitis CM, Lease ED, Edelman J, Kirby KA, Leisenring WM, et al. Symptomatic Respiratory Virus Infection and Chronic Lung Allograft Dysfunction. Clin Infect Dis. 2016;62(3):313–9. PubMed Central PMCID: PMCPMC4706632. doi: 10.1093/cid/civ871 - DOI - PMC - PubMed
-
- Fishman JA. Infection in solid-organ transplant recipients. N Engl J Med. 2007;357(25):2601–14. doi: 10.1056/NEJMra064928 - DOI - PubMed
-
- Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, et al. A new arenavirus in a cluster of fatal transplant-associated diseases. The New England journal of medicine. 2008;358(10):991–8. doi: 10.1056/NEJMoa073785 - DOI - PubMed
-
- Wunderli W, Meerbach A, Guengoer T, Berger C, Greiner O, Caduff R, et al. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PloS one. 2011;6(11):e27483 doi: 10.1371/journal.pone.0027483 - DOI - PMC - PubMed
-
- Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerging Infectious Diseases. 2010;16(6):918–25. doi: 10.3201/eid1606.091536 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

