Identification and expression analysis of starch branching enzymes involved in starch synthesis during the development of chestnut (Castanea mollissima Blume) cotyledons
- PMID: 28542293
- PMCID: PMC5441625
- DOI: 10.1371/journal.pone.0177792
Identification and expression analysis of starch branching enzymes involved in starch synthesis during the development of chestnut (Castanea mollissima Blume) cotyledons
Abstract
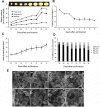
Chinese chestnut (Castanea mollissima Blume) is native to China and distributes widely in arid and semi-arid mountain area with barren soil. As a perennial crop, chestnut is an alternative food source and acts as an important commercial nut tree in China. Starch is the major metabolite in nuts, accounting for 46 ~ 64% of the chestnut dry weight. The accumulation of total starch and amylopectin showed a similar increasing trend during the development of nut. Amylopectin contributed up to 76% of the total starch content at 80 days after pollination (DAP). The increase of total starch mainly results from amylopectin synthesis. Among genes associated with starch biosynthesis, CmSBEs (starch branching enzyme) showed significant increase during nut development. Two starch branching enzyme isoforms, CmSBE I and CmSBE II, were identified from chestnut cotyledon using zymogram analysis. CmSBE I and CmSBE II showed similar patterns of expression during nut development. The accumulations of CmSBE transcripts and proteins in developing cotyledons were characterized. The expressions of two CmSBE genes increased from 64 DAP and reached the highest levels at 77 DAP, and SBE activity reached its peak at 74 DAP. These results suggested that the CmSBE enzymes mainly contributed to amylopectin synthesis and influenced the amylopectin content in the developing cotyledon, which would be beneficial to chestnut germplasm selection and breeding.
Conflict of interest statement
Figures



Similar articles
-
Transcriptomic identification and expression of starch and sucrose metabolism genes in the seeds of Chinese chestnut (Castanea mollissima).J Agric Food Chem. 2015 Jan 28;63(3):929-42. doi: 10.1021/jf505247d. Epub 2015 Jan 13. J Agric Food Chem. 2015. PMID: 25537355
-
Signatures of Selection in the Genomes of Chinese Chestnut (Castanea mollissima Blume): The Roots of Nut Tree Domestication.Front Plant Sci. 2018 Jun 25;9:810. doi: 10.3389/fpls.2018.00810. eCollection 2018. Front Plant Sci. 2018. PMID: 29988533 Free PMC article.
-
Roles of the GA-mediated SPL Gene Family and miR156 in the Floral Development of Chinese Chestnut (Castanea mollissima).Int J Mol Sci. 2019 Mar 29;20(7):1577. doi: 10.3390/ijms20071577. Int J Mol Sci. 2019. PMID: 30934840 Free PMC article.
-
Progress in controlling starch structure by modifying starch-branching enzymes.Planta. 2016 Jan;243(1):13-22. doi: 10.1007/s00425-015-2421-2. Planta. 2016. PMID: 26486516 Review.
-
A review of starch-branching enzymes and their role in amylopectin biosynthesis.IUBMB Life. 2014 Aug;66(8):546-58. doi: 10.1002/iub.1297. Epub 2014 Sep 5. IUBMB Life. 2014. PMID: 25196474 Review.
Cited by
-
Utilization of Germinated Seeds as Functional Food Ingredients: Optimization of Nutrient Composition and Antioxidant Activity Evolution Based on the Germination Characteristics of Chinese Chestnut (Castanea mollissima).Foods. 2024 Aug 20;13(16):2605. doi: 10.3390/foods13162605. Foods. 2024. PMID: 39200532 Free PMC article.
-
Beta-Amylase and Phosphatidic Acid Involved in Recalcitrant Seed Germination of Chinese Chestnut.Front Plant Sci. 2022 Mar 25;13:828270. doi: 10.3389/fpls.2022.828270. eCollection 2022. Front Plant Sci. 2022. PMID: 35401618 Free PMC article.
-
Comparison of Physicochemical Properties of Starches from Nine Chinese Chestnut Varieties.Molecules. 2018 Dec 7;23(12):3248. doi: 10.3390/molecules23123248. Molecules. 2018. PMID: 30544638 Free PMC article.
-
Digital Absolute Gene Expression Analysis of Essential Starch-Related Genes in a Radiation Developed Amaranthus cruentus L. Variety in Comparison with Real-Time PCR.Plants (Basel). 2020 Jul 30;9(8):966. doi: 10.3390/plants9080966. Plants (Basel). 2020. PMID: 32751665 Free PMC article.
-
Molecular identification of the key starch branching enzyme-encoding gene SBE2.3 and its interacting transcription factors in banana fruits.Hortic Res. 2020 Jul 1;7:101. doi: 10.1038/s41438-020-0325-1. eCollection 2020. Hortic Res. 2020. PMID: 32637129 Free PMC article.
References
-
- Smith AM, Denyer K, Martin C. The synthesis of the starch granule. Annu Rev Plant Physiol Plant Mol Biol. 1997; 48: 67–87. doi: 10.1146/annurev.arplant.48.1.67 - DOI - PubMed
-
- Diego F, Kathleen GH, Shelley J. Starch characteristics of modern and heirloom potato cultivars. Am J Potato Res. 2013;
-
- Botticella E, Sestili F, Lafiandra D. Characterization of SBEIIa homoeologous genes in bread wheat. Mol Genet Genomics. 2012; 287: 515–524. doi: 10.1007/s00438-012-0694-8 - DOI - PubMed
-
- Jiang L, Yu X, Qi X, Yu Q, Deng S, Bai B, et al. Multigene engineering of starch biosynthesis in maize endosperm increases the total starch content and the proportion of amylose. Transgenic Res. 2013; 22, 1133–1142. doi: 10.1007/s11248-013-9717-4 - DOI - PubMed
-
- Syahariza ZA, Sar S, Hasjim J, Tizzotti MJ, Gilbert RG. The importance of amylose and amylopectin fine structures for starch digestibility in cooked rice grains. Food Chem. 2013; 136: 742–749. doi: 10.1016/j.foodchem.2012.08.053 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

