Genome-scale investigation of olfactory system spatial heterogeneity
- PMID: 28542411
- PMCID: PMC5443560
- DOI: 10.1371/journal.pone.0178087
Genome-scale investigation of olfactory system spatial heterogeneity
Abstract
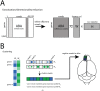
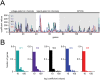
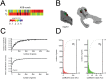
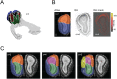
The early olfactory system is organized in parallel, with numerous, specialized subsystems established by the modular and topographic projections of sensory inputs. While these anatomical sub-systems are in many cases demarcated by well-known marker genes, we stand to learn considerably more about their possible functional specializations from comprehensive, genome-scale descriptions of their molecular anatomy. Here, we leverage the resources of the Allen Brain Atlas (ABA)-a spatially registered compendium of gene expression for the mouse brain-to investigate the early olfactory system's genomic anatomy. We cluster thousands of genes across thousands of voxels in the ABA to derive several novel parcellations of the olfactory system, and concomitantly discover novel sets of enriched, subregion-specific genes that can serve as a starting point for future inquiry.
Conflict of interest statement
Figures



References
-
- Plato, Helmbold WC, Rabninowitz W. Phaedrus. Indianapolis: Bobbs-Merrill Co.; 1956.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

