Genet-specific DNA methylation probabilities detected in a spatial epigenetic analysis of a clonal plant population
- PMID: 28542457
- PMCID: PMC5439711
- DOI: 10.1371/journal.pone.0178145
Genet-specific DNA methylation probabilities detected in a spatial epigenetic analysis of a clonal plant population
Abstract
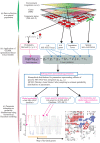
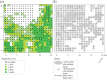
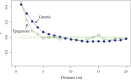
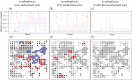
In sessile organisms such as plants, spatial genetic structures of populations show long-lasting patterns. These structures have been analyzed across diverse taxa to understand the processes that determine the genetic makeup of organismal populations. For many sessile organisms that mainly propagate via clonal spread, epigenetic status can vary between clonal individuals in the absence of genetic changes. However, fewer previous studies have explored the epigenetic properties in comparison to the genetic properties of natural plant populations. Here, we report the simultaneous evaluation of the spatial structure of genetic and epigenetic variation in a natural population of the clonal plant Cardamine leucantha. We applied a hierarchical Bayesian model to evaluate the effects of membership of a genet (a group of individuals clonally derived from a single seed) and vegetation cover on the epigenetic variation between ramets (clonal plants that are physiologically independent individuals). We sampled 332 ramets in a 20 m × 20 m study plot that contained 137 genets (identified using eight SSR markers). We detected epigenetic variation in DNA methylation at 24 methylation-sensitive amplified fragment length polymorphism (MS-AFLP) loci. There were significant genet effects at all 24 MS-AFLP loci in the distribution of subepiloci. Vegetation cover had no statistically significant effect on variation in the majority of MS-AFLP loci. The spatial aggregation of epigenetic variation is therefore largely explained by the aggregation of ramets that belong to the same genets. By applying hierarchical Bayesian analyses, we successfully identified a number of genet-specific changes in epigenetic status within a natural plant population in a complex context, where genotypes and environmental factors are unevenly distributed. This finding suggests that it requires further studies on the spatial epigenetic structure of natural populations of diverse organisms, particularly for sessile clonal species.
Conflict of interest statement
Figures


References
-
- Jackson JBC, Buss LW, Cook RE. Population biology and evolution of clonal organisms. New Haven: Yale University; 1985.
-
- van Groenendael JM, de Kroon H. Clonal growth in plants: regulation and function. The Hague: SPB Academic Publishing; 1990.
-
- de Kroon H, Hutchings MJ. Morphological plasticity in clonal plants: the foraging concept reconsidered. J Ecol. 1995; 83: 143–152.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

