A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer
- PMID: 28542458
- PMCID: PMC5444587
- DOI: 10.1371/journal.pone.0177062
A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer
Abstract
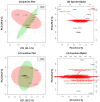
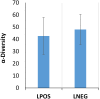
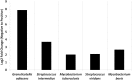
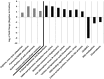
Lung cancer (LC) is the most prevalent cancer worldwide, and responsible for over 1.3 million deaths each year. Currently, LC has a low five year survival rates relative to other cancers, and thus, novel methods to screen for and diagnose malignancies are necessary to improve patient outcomes. Here, we report on a pilot-sized study to evaluate the potential of the sputum microbiome as a source of non-invasive bacterial biomarkers for lung cancer status and stage. Spontaneous sputum samples were collected from ten patients referred with possible LC, of which four were eventually diagnosed with LC (LC+), and six had no LC after one year (LC-). Of the seven bacterial species found in all samples, Streptococcus viridans was significantly higher in LC+ samples. Seven further bacterial species were found only in LC-, and 16 were found only in samples from LC+. Additional taxonomic differences were identified in regards to significant fold changes between LC+ and LC-cases, with five species having significantly higher abundances in LC+, with Granulicatella adiacens showing the highest level of abundance change. Functional differences, evident through significant fold changes, included polyamine metabolism and iron siderophore receptors. G. adiacens abundance was correlated with six other bacterial species, namely Enterococcus sp. 130, Streptococcus intermedius, Escherichia coli, S. viridans, Acinetobacter junii, and Streptococcus sp. 6, in LC+ samples only, which could also be related to LC stage. Spontaneous sputum appears to be a viable source of bacterial biomarkers which may have utility as biomarkers for LC status and stage.
Conflict of interest statement
Figures
Similar articles
-
Taxonomic diversity of sputum microbiome in lung cancer patients and its relationship with chromosomal aberrations in blood lymphocytes.Sci Rep. 2020 Jun 15;10(1):9681. doi: 10.1038/s41598-020-66654-x. Sci Rep. 2020. PMID: 32541778 Free PMC article.
-
The metabolomic detection of lung cancer biomarkers in sputum.Lung Cancer. 2016 Apr;94:88-95. doi: 10.1016/j.lungcan.2016.02.006. Epub 2016 Feb 8. Lung Cancer. 2016. PMID: 26973212
-
Genetic damage in lymphocytes of lung cancer patients is correlated to the composition of the respiratory tract microbiome.Mutagenesis. 2021 May 31;36(2):143-153. doi: 10.1093/mutage/geab004. Mutagenesis. 2021. PMID: 33454779
-
Lung cancer: a new frontier for microbiome research and clinical translation.Ecancermedicalscience. 2018 Sep 5;12:866. doi: 10.3332/ecancer.2018.866. eCollection 2018. Ecancermedicalscience. 2018. PMID: 30263057 Free PMC article. Review.
-
Microbiome Dysbiosis and Predominant Bacterial Species as Human Cancer Biomarkers.J Gastrointest Cancer. 2020 Sep;51(3):725-728. doi: 10.1007/s12029-019-00311-z. J Gastrointest Cancer. 2020. PMID: 31605288 Review.
Cited by
-
Microbial Biomarkers for Lung Cancer: Current Understandings and Limitations.J Clin Med. 2022 Dec 8;11(24):7298. doi: 10.3390/jcm11247298. J Clin Med. 2022. PMID: 36555915 Free PMC article. Review.
-
Alterations of fecal bacterial communities in patients with lung cancer.Am J Transl Res. 2018 Oct 15;10(10):3171-3185. eCollection 2018. Am J Transl Res. 2018. PMID: 30416659 Free PMC article.
-
PARP1 Is Up-Regulated in Non-small Cell Lung Cancer Tissues in the Presence of the Cyanobacterial Toxin Microcystin.Front Microbiol. 2018 Aug 6;9:1757. doi: 10.3389/fmicb.2018.01757. eCollection 2018. Front Microbiol. 2018. PMID: 30127774 Free PMC article.
-
Sputum Microbiota in Coal Workers Diagnosed with Pneumoconiosis as Revealed by 16S rRNA Gene Sequencing.Life (Basel). 2022 Jun 2;12(6):830. doi: 10.3390/life12060830. Life (Basel). 2022. PMID: 35743861 Free PMC article.
-
Analysis of Gut Microbiota Signature and Microbe-Disease Progression Associations in Locally Advanced Non-Small Cell Lung Cancer Patients Treated With Concurrent Chemoradiotherapy.Front Cell Infect Microbiol. 2022 Jun 1;12:892401. doi: 10.3389/fcimb.2022.892401. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35719339 Free PMC article.
References
-
- Who. Cancer Factsheet. WHO Fact Sheets, Number 2970. 2013.
-
- Jemal A, Siegel R, Xu JQ, Ward E. Cancer Statistics, 2010. CA: A Cancer Journal for Clinicians. 2010;60(5):277–300. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

