The PET and LIM1-2 domains of testin contribute to intramolecular and homodimeric interactions
- PMID: 28542564
- PMCID: PMC5436826
- DOI: 10.1371/journal.pone.0177879
The PET and LIM1-2 domains of testin contribute to intramolecular and homodimeric interactions
Abstract
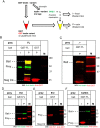
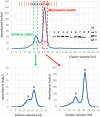
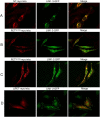
The focal adhesion protein testin is a modular scaffold and tumour suppressor that consists of an N-terminal cysteine rich (CR) domain, a PET domain of unknown function and three C-terminal LIM domains. Testin has been proposed to have an open and a closed conformation based on the observation that its N-terminal half and C-terminal half directly interact. Here we extend the testin conformational model by demonstrating that testin can also form an antiparallel homodimer. In support of this extended model we determined that the testin region (amino acids 52-233) harbouring the PET domain interacts with the C-terminal LIM1-2 domains in vitro and in cells, and assign a critical role to tyrosine 288 in this interaction.
Conflict of interest statement
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

