Regulation of acetyl-CoA synthetase transcription by the CrbS/R two-component system is conserved in genetically diverse environmental pathogens
- PMID: 28542616
- PMCID: PMC5436829
- DOI: 10.1371/journal.pone.0177825
Regulation of acetyl-CoA synthetase transcription by the CrbS/R two-component system is conserved in genetically diverse environmental pathogens
Abstract
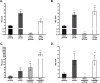
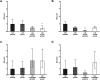
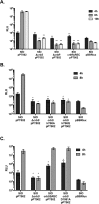
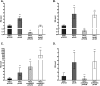
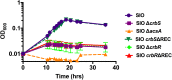
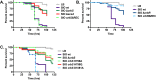
The CrbS/R two-component signal transduction system is a conserved regulatory mechanism through which specific Gram-negative bacteria control acetate flux into primary metabolic pathways. CrbS/R governs expression of acetyl-CoA synthase (acsA), an enzyme that converts acetate to acetyl-CoA, a metabolite at the nexus of the cell's most important energy-harvesting and biosynthetic reactions. During infection, bacteria can utilize this system to hijack host acetate metabolism and alter the course of colonization and pathogenesis. In toxigenic strains of Vibrio cholerae, CrbS/R-dependent expression of acsA is required for virulence in an arthropod model. Here, we investigate the function of the CrbS/R system in Pseudomonas aeruginosa, Pseudomonas entomophila, and non-toxigenic V. cholerae strains. We demonstrate that its role in acetate metabolism is conserved; this system regulates expression of the acsA gene and is required for growth on acetate as a sole carbon source. As a first step towards describing the mechanism of signaling through this pathway, we identify residues and domains that may be critical for phosphotransfer. We further demonstrate that although CrbS, the putative hybrid sensor kinase, carries both a histidine kinase domain and a receiver domain, the latter is not required for acsA transcription. In order to determine whether our findings are relevant to pathogenesis, we tested our strains in a Drosophila model of oral infection previously employed for the study of acetate-dependent virulence by V. cholerae. We show that non-toxigenic V. cholerae strains lacking CrbS or CrbR are significantly less virulent than are wild-type strains, while P. aeruginosa and P. entomophila lacking CrbS or CrbR are fully pathogenic. Together, the data suggest that the CrbS/R system plays a central role in acetate metabolism in V. cholerae, P. aeruginosa, and P. entomophila. However, each microbe's unique environmental adaptations and pathogenesis strategies may dictate conditions under which CrbS/R-mediated acs expression is most critical.
Conflict of interest statement
Figures




Similar articles
-
Modulation of CrbS-Dependent Activation of the Acetate Switch in Vibrio cholerae.J Bacteriol. 2018 Nov 6;200(23):e00380-18. doi: 10.1128/JB.00380-18. Print 2018 Dec 1. J Bacteriol. 2018. PMID: 30224439 Free PMC article.
-
A Putative Acetylation System in Vibrio cholerae Modulates Virulence in Arthropod Hosts.Appl Environ Microbiol. 2018 Oct 17;84(21):e01113-18. doi: 10.1128/AEM.01113-18. Print 2018 Nov 1. Appl Environ Microbiol. 2018. PMID: 30143508 Free PMC article.
-
Characterization of the CrbS/R Two-Component System in Pseudomonas fluorescens Reveals a New Set of Genes under Its Control and a DNA Motif Required for CrbR-Mediated Transcriptional Activation.Front Microbiol. 2017 Nov 20;8:2287. doi: 10.3389/fmicb.2017.02287. eCollection 2017. Front Microbiol. 2017. PMID: 29250042 Free PMC article.
-
Expression of Vibrio cholerae virulence genes in response to environmental signals.Curr Issues Intest Microbiol. 2002 Sep;3(2):29-38. Curr Issues Intest Microbiol. 2002. PMID: 12400636 Review.
-
Genetics of stress adaptation and virulence in toxigenic Vibrio cholerae.DNA Cell Biol. 2004 Nov;23(11):723-41. doi: 10.1089/dna.2004.23.723. DNA Cell Biol. 2004. PMID: 15585131 Review.
Cited by
-
Bacterial acetate metabolism and its influence on human epithelia.Emerg Top Life Sci. 2024 Feb 22;8(1):1-13. doi: 10.1042/ETLS20220092. Emerg Top Life Sci. 2024. PMID: 36945843 Free PMC article. Review.
-
Metabolic engineering of Pseudomonas taiwanensis VLB120 for rhamnolipid biosynthesis from biomass-derived aromatics.Metab Eng Commun. 2022 Aug 10;15:e00202. doi: 10.1016/j.mec.2022.e00202. eCollection 2022 Dec. Metab Eng Commun. 2022. PMID: 36017490 Free PMC article.
-
Involvement of the MxtR/ErdR (CrbS/CrbR) Two-Component System in Acetate Metabolism in Pseudomonas putida KT2440.Microorganisms. 2021 Jul 22;9(8):1558. doi: 10.3390/microorganisms9081558. Microorganisms. 2021. PMID: 34442637 Free PMC article.
-
Contribution of Uncharacterized Target Genes of MxtR/ErdR to Carbon Source Utilization by Pseudomonas putida KT2440.Microbiol Spectr. 2023 Feb 14;11(1):e0292322. doi: 10.1128/spectrum.02923-22. Epub 2022 Dec 13. Microbiol Spectr. 2023. PMID: 36511656 Free PMC article.
-
The Regulatory Hierarchy Following Signal Integration by the CbrAB Two-Component System: Diversity of Responses and Functions.Genes (Basel). 2022 Feb 18;13(2):375. doi: 10.3390/genes13020375. Genes (Basel). 2022. PMID: 35205417 Free PMC article. Review.
References
-
- Tzou P, De Gregorio E, Lemaitre B. How Drosophila combats microbial infection: a model to study innate immunity and host–pathogen interactions. Curr Opin Microbiol. 2002;5: 102–110. - PubMed
-
- Panayidou S, Ioannidou E, Apidianakis Y. Human pathogenic bacteria, fungi, and viruses in Drosophila. Virulence. 2014;5: 253–269. doi: 10.4161/viru.27524 - DOI - PMC - PubMed
-
- Fauvarque M-O. Small flies to tackle big questions: assaying complex bacterial virulence mechanisms using Drosophila melanogaster. Cell Microbiol. 2014;16: 824–833. doi: 10.1111/cmi.12292 - DOI - PubMed
-
- Vodovar N, Acosta C, Lemaitre B, Boccard F. Drosophila: a polyvalent model to decipher host–pathogen interactions. Trends Microbiol. 2004;12: 235–242. doi: 10.1016/j.tim.2004.03.007 - DOI - PubMed
-
- Vallet-Gely I, Opota O, Boniface A, Novikov A, Lemaitre B. A secondary metabolite acting as a signalling molecule controls Pseudomonas entomophila virulence. Cell Microbiol. 2010;12: 1666–1679. doi: 10.1111/j.1462-5822.2010.01501.x - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

