Mechanism of graphene-induced cytotoxicity: Role of endonucleases
- PMID: 28543094
- PMCID: PMC12448367
- DOI: 10.1002/jat.3462
Mechanism of graphene-induced cytotoxicity: Role of endonucleases
Abstract
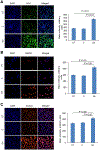
Graphene, a crystalline allotrope or carbon, presents numerous useful properties; however, its toxicity is yet to be determined. One of the most dramatic and irreversible toxic abilities of carbon nanomaterials is the induction of DNA fragmentation produced by endogenous cellular endonucleases. This study demonstrated that pristine graphene exposed to cultured kidney tubular epithelial cells is capable of inducing DNA fragmentation measured by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, which is usually associated with cell death. TUNEL (cell death) and endonuclease activity measured using a near infrared fluorescence probe was significantly higher in cells containing graphene aggregates detected by Raman spectroscopy. The elevation of TUNEL coincided with the increased abundance of heme oxygenase 1 (HO-1), heat shock protein 90 (HSP90), active caspase-3 and endonucleases (deoxyribonuclease I [DNase I] and endonuclease G [EndoG]), as measured by quantitative immunocytochemistry. Specific inhibitors for HO-1, HSP90, caspase-3, DNase I and EndoG almost completely blocked the DNA fragmentation induced by graphene exposure. Therefore, graphene induces cell death through oxidative injury, caspase-mediated and caspase-independent pathways; and endonucleases DNase I and EndoG are important for graphene toxicity. Inhibition of these pathways may ameliorate cell injury produced by graphene. Copyright © 2017 John Wiley & Sons, Ltd.
Keywords: DNA fragmentation; DNase I; EndoG; cell death; endonucleases; graphene; oxidative injury.
Copyright © 2017 John Wiley & Sons, Ltd.
Conflict of interest statement
Conflict of interest
The authors did not report any conflict of interest.
Figures



References
-
- Ahamed M, Alsalhi MS, Siddiqui MK. 2010. Silver nanoparticle applications and human health. Clin. Chim. Acta 411: 1841–1848. - PubMed
-
- Akhavan O, Ghaderi E. 2010. Toxicity of graphene and graphene oxide nanowalls against bacteria. ACS Nano 4: 5731–5736. - PubMed
-
- Basnakian AG, Kaushal GP, Shah SV. 2002. Apoptotic pathways of oxidative damage to renal tubular epithelial cells. Antioxid. Redox Signal 4: 915–924. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

