The best models of metabolism
- PMID: 28544810
- PMCID: PMC5643013
- DOI: 10.1002/wsbm.1391
The best models of metabolism
Abstract
Biochemical systems are among of the oldest application areas of mathematical modeling. Spanning a time period of over one hundred years, the repertoire of options for structuring a model and for formulating reactions has been constantly growing, and yet, it is still unclear whether or to what degree some models are better than others and how the modeler is to choose among them. In fact, the variety of options has become overwhelming and difficult to maneuver for novices and experts alike. This review outlines the metabolic model design process and discusses the numerous choices for modeling frameworks and mathematical representations. It tries to be inclusive, even though it cannot be complete, and introduces the various modeling options in a manner that is as unbiased as that is feasible. However, the review does end with personal recommendations for the choices of default models. WIREs Syst Biol Med 2017, 9:e1391. doi: 10.1002/wsbm.1391 For further resources related to this article, please visit the WIREs website.
© 2017 Wiley Periodicals, Inc.
Conflict of interest statement
The author declares no conflicts of interest.
Figures
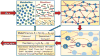
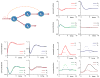
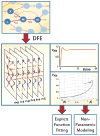
Similar articles
-
Computational modeling approaches to the dynamics of oncolytic viruses.Wiley Interdiscip Rev Syst Biol Med. 2016 May;8(3):242-52. doi: 10.1002/wsbm.1332. Epub 2016 Mar 22. Wiley Interdiscip Rev Syst Biol Med. 2016. PMID: 27001049 Free PMC article. Review.
-
Mathematical modeling of cardiac growth and remodeling.Wiley Interdiscip Rev Syst Biol Med. 2016 May;8(3):211-26. doi: 10.1002/wsbm.1330. Epub 2016 Mar 7. Wiley Interdiscip Rev Syst Biol Med. 2016. PMID: 26952285 Free PMC article. Review.
-
Genome-scale metabolic models applied to human health and disease.Wiley Interdiscip Rev Syst Biol Med. 2017 Nov;9(6). doi: 10.1002/wsbm.1393. Epub 2017 Jun 23. Wiley Interdiscip Rev Syst Biol Med. 2017. PMID: 28644920 Review.
-
Modeling, signaling and cytoskeleton dynamics: integrated modeling-experimental frameworks in cell migration.Wiley Interdiscip Rev Syst Biol Med. 2017 Jan;9(1):10.1002/wsbm.1365. doi: 10.1002/wsbm.1365. Epub 2016 Nov 15. Wiley Interdiscip Rev Syst Biol Med. 2017. PMID: 27863122 Free PMC article. Review.
-
Understanding the mTOR signaling pathway via mathematical modeling.Wiley Interdiscip Rev Syst Biol Med. 2017 Jul;9(4):e1379. doi: 10.1002/wsbm.1379. Epub 2017 Feb 10. Wiley Interdiscip Rev Syst Biol Med. 2017. PMID: 28186392 Free PMC article. Review.
Cited by
-
Ensemble methods for stochastic networks with special reference to the biological clock of Neurospora crassa.PLoS One. 2018 May 16;13(5):e0196435. doi: 10.1371/journal.pone.0196435. eCollection 2018. PLoS One. 2018. PMID: 29768444 Free PMC article.
-
GEMCAT-a new algorithm for gene expression-based prediction of metabolic alterations.NAR Genom Bioinform. 2025 Jan 31;7(1):lqaf003. doi: 10.1093/nargab/lqaf003. eCollection 2025 Mar. NAR Genom Bioinform. 2025. PMID: 39897103 Free PMC article.
-
What's next for computational systems biology?Front Syst Biol. 2023 Sep 19;3:1250228. doi: 10.3389/fsysb.2023.1250228. eCollection 2023. Front Syst Biol. 2023. PMID: 40809471 Free PMC article. Review.
-
A model of dopamine and serotonin-kynurenine metabolism in cortisolemia: Implications for depression.PLoS Comput Biol. 2021 May 10;17(5):e1008956. doi: 10.1371/journal.pcbi.1008956. eCollection 2021 May. PLoS Comput Biol. 2021. PMID: 33970902 Free PMC article.
-
Optimal control of agent-based models via surrogate modeling.PLoS Comput Biol. 2025 Jan 14;21(1):e1012138. doi: 10.1371/journal.pcbi.1012138. eCollection 2025 Jan. PLoS Comput Biol. 2025. PMID: 39808665 Free PMC article.
References
-
- Michaelis L, Menten ML. Die Kinetik der Invertinwirkung. Biochemische Zeitschrift. 1913;49:333–369.
-
- Henri MV. Lois générales de l’action des diastases. Paris: Hermann; 1903.
-
- Savageau MA. Enzyme kinetics in vitro and in vivo: Michaelis-Menten revisited. In: Bittar EE, editor. Principles of Medical Biology. Vol. 4. Greenwich, CT: JAI Press Inc; 1995. pp. 93–146.
-
- Tummler K, Lubitz T, Schelker M, Klipp E. New types of experimental data shape the use of enzyme kinetics for dynamic network modeling. FEBS J. 2014;281:549–571. - PubMed
-
- Voit EO, Sands PJ. Modeling forest growth. I. Canonical approach. Ecol Modeling. 1996;86:51–71.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

