An unusual intragenic promoter of PIWIL2 contributes to aberrant activation of oncogenic PL2L60
- PMID: 28545024
- PMCID: PMC5542253
- DOI: 10.18632/oncotarget.17553
An unusual intragenic promoter of PIWIL2 contributes to aberrant activation of oncogenic PL2L60
Abstract
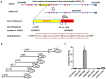
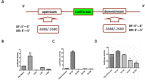
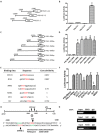
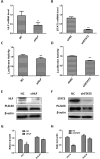
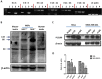
PIWIL2-like (PL2L) protein 60 (PL2L60), a product of aberrantly activated PIWIL2 gene, is widely expressed in various types of tumors and may promote tumorigenesis. However, the mechanisms underlying the activation of expression of PL2L60 remain unknown. In this study, an intragenic promoter responsible for the activation of PL2L60 within the human PIWIL2 gene has been identified, cloned and characterized. The promoter of PL2L60 is located in the intron 10 of the host gene PIWIL2. Bioinformatic and mutagenic analysis reveals that this intragenic promoter within the sequence of 50 nucleotides contains two closely arranged cis-acting elements specific for the hepatic leukemia factor (HLF) in the positive strand and signal transducer and activator of transcription 3 (STAT3) in the negative strand. Chromatin immunoprecipitation analysis demonstrates that both the HLF and polymerase II (Pol II), a hallmark of active promoters, directly bind to the sequence, although STAT3 does not. Knockdown of HLF and STAT3 alone or both by RNA interference significantly reduced both promoter activity and the PL2L60 protein expression, although there is no additive effect. The expression of PL2L60 proteins was enhanced when host gene Piwil2 was genetically disrupted in a murine cell model. Taken together, we have identified a PL2L60-specific intragenic promoter in the host gene of PIWIL2, which is interdependently activated by HLF and STAT3 through steric interaction. This activation is dependent on cellular milieu rather than the integrity of host gene PIWIL2, highlighting a novel, important mechanism for a cancer-causing gene to be activated during tumorigenesis.
Keywords: PIWIL2; PL2L60; STAT3; alienated gene activation; intragenic promoter.
Figures

References
-
- Gao JX, Liu N, Wu HL. PIWIL2 (piwi-like RNA-mediated gene silencing 2) Atlas Genet Cytogenet Oncol Haematol. 2014;18:919–927.
-
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. - PubMed
-
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–133. - PubMed
-
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

