Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes
- PMID: 28545061
- PMCID: PMC5448818
- DOI: 10.1371/journal.ppat.1006391
Microbiota-induced peritrophic matrix regulates midgut homeostasis and prevents systemic infection of malaria vector mosquitoes
Abstract
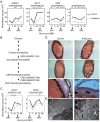
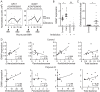
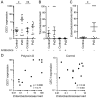
Manipulation of the mosquito gut microbiota can lay the foundations for novel methods for disease transmission control. Mosquito blood feeding triggers a significant, transient increase of the gut microbiota, but little is known about the mechanisms by which the mosquito controls this bacterial growth whilst limiting inflammation of the gut epithelium. Here, we investigate the gut epithelial response to the changing microbiota load upon blood feeding in the malaria vector Anopheles coluzzii. We show that the synthesis and integrity of the peritrophic matrix, which physically separates the gut epithelium from its luminal contents, is microbiota dependent. We reveal that the peritrophic matrix limits the growth and persistence of Enterobacteriaceae within the gut, whilst preventing seeding of a systemic infection. Our results demonstrate that the peritrophic matrix is a key regulator of mosquito gut homeostasis and establish functional analogies between this and the mucus layers of the mammalian gastrointestinal tract.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Immune resistance and tolerance strategies in malaria vector and non-vector mosquitoes.Parasit Vectors. 2017 Apr 18;10(1):186. doi: 10.1186/s13071-017-2109-5. Parasit Vectors. 2017. PMID: 28420446 Free PMC article.
-
PGRP-LD mediates A. stephensi vector competency by regulating homeostasis of microbiota-induced peritrophic matrix synthesis.PLoS Pathog. 2018 Feb 28;14(2):e1006899. doi: 10.1371/journal.ppat.1006899. eCollection 2018 Feb. PLoS Pathog. 2018. PMID: 29489896 Free PMC article.
-
Gut microbiota is essential in PGRP-LA regulated immune protection against Plasmodium berghei infection.Parasit Vectors. 2020 Jan 6;13(1):3. doi: 10.1186/s13071-019-3876-y. Parasit Vectors. 2020. PMID: 31907025 Free PMC article.
-
The tripartite interactions between the mosquito, its microbiota and Plasmodium.Parasit Vectors. 2018 Mar 20;11(1):200. doi: 10.1186/s13071-018-2784-x. Parasit Vectors. 2018. PMID: 29558973 Free PMC article. Review.
-
The microbiota, the malarial parasite, and the mosquito [MMM] - A three-sided relationship.Mol Biochem Parasitol. 2023 Feb;253:111543. doi: 10.1016/j.molbiopara.2023.111543. Epub 2023 Jan 13. Mol Biochem Parasitol. 2023. PMID: 36642385 Review.
Cited by
-
Composition and functional roles of the gut microbiota in mosquitoes.Curr Opin Insect Sci. 2018 Aug;28:59-65. doi: 10.1016/j.cois.2018.05.008. Epub 2018 May 22. Curr Opin Insect Sci. 2018. PMID: 30551768 Free PMC article. Review.
-
The mosquito holobiont: fresh insight into mosquito-microbiota interactions.Microbiome. 2018 Mar 20;6(1):49. doi: 10.1186/s40168-018-0435-2. Microbiome. 2018. PMID: 29554951 Free PMC article. Review.
-
Ironing out the Details: Exploring the Role of Iron and Heme in Blood-Sucking Arthropods.Front Physiol. 2018 Jan 17;8:1134. doi: 10.3389/fphys.2017.01134. eCollection 2017. Front Physiol. 2018. PMID: 29387018 Free PMC article. Review.
-
Impact of Gut Bacteria on the Infection and Transmission of Pathogenic Arboviruses by Biting Midges and Mosquitoes.Microb Ecol. 2020 Oct;80(3):703-717. doi: 10.1007/s00248-020-01517-6. Epub 2020 May 27. Microb Ecol. 2020. PMID: 32462391 Free PMC article.
-
Impact of insecticide resistance on malaria vector competence: a literature review.Malar J. 2023 Jan 17;22(1):19. doi: 10.1186/s12936-023-04444-2. Malar J. 2023. PMID: 36650503 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

