Pre-silencing of genes involved in the electron transport chain (ETC) pathway is associated with responsiveness to abatacept in rheumatoid arthritis
- PMID: 28545499
- PMCID: PMC5445375
- DOI: 10.1186/s13075-017-1319-8
Pre-silencing of genes involved in the electron transport chain (ETC) pathway is associated with responsiveness to abatacept in rheumatoid arthritis
Abstract
Background: In the current context of personalized medicine, one of the major challenges in the management of rheumatoid arthritis (RA) is to identify biomarkers that predict drug responsiveness. From the European APPRAISE trial, our main objective was to identify a gene expression profile associated with responsiveness to abatacept (ABA) + methotrexate (MTX) and to understand the involvement of this signature in the pathophysiology of RA.
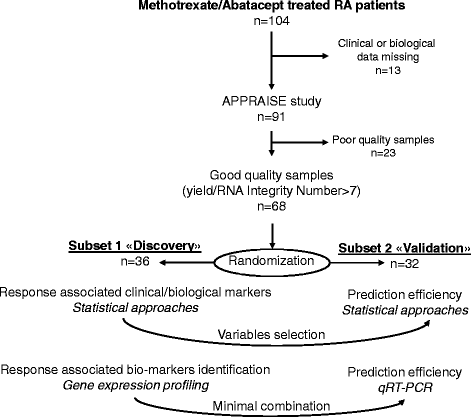
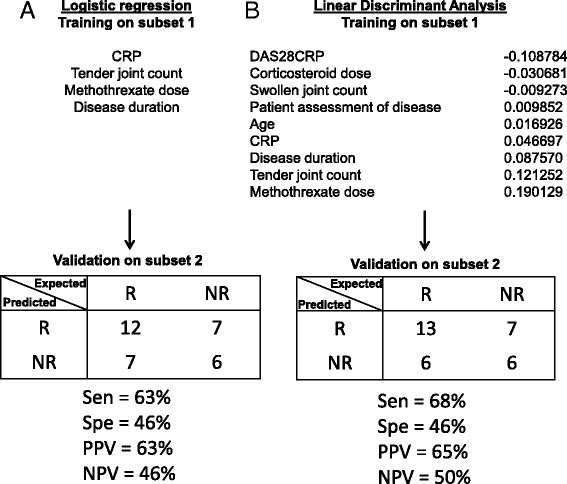
Methods: Whole human genome microarrays (4 × 44 K) were performed from a first subset of 36 patients with RA. Data validation by quantitative reverse-transcription (qRT)-PCR was performed from a second independent subset of 32 patients with RA. Gene Ontology and WikiPathways database allowed us to highlight the specific biological mechanisms involved in predicting response to ABA/MTX.
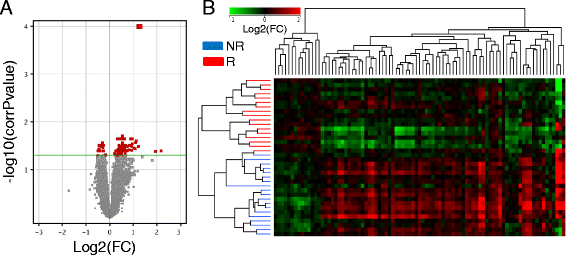
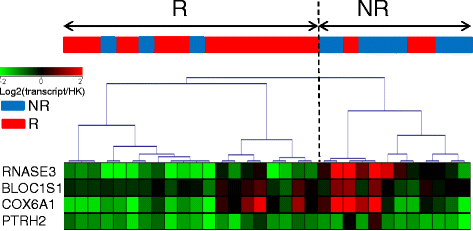
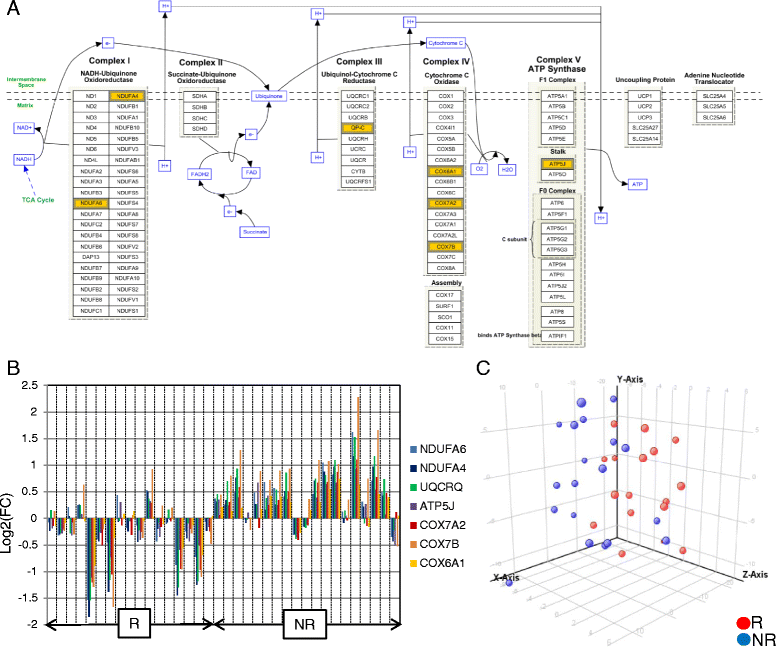
Results: From the first subset of 36 patients with RA, a combination including 87 transcripts allowed almost perfect separation between responders and non-responders to ABA/MTX. Next, the second subset of patients 32 with RA allowed validation by qRT-PCR of a minimal signature with only four genes. This latter signature categorized 81% of patients with RA with 75% sensitivity, 85% specificity and 85% negative predictive value. This combination showed a significant enrichment of genes involved in electron transport chain (ETC) pathways. Seven transcripts from ETC pathways (NDUFA6, NDUFA4, UQCRQ, ATP5J, COX7A2, COX7B, COX6A1) were significantly downregulated in responders versus non-responders to ABA/MTX. Moreover, dysregulation of these genes was independent of inflammation and was specific to ABA response.
Conclusion: Pre-silencing of ETC genes is associated with future response to ABA/MTX and might be a crucial key to susceptibility to ABA response.
Keywords: Abatacept; Abatacept response; Biomarkers; Microarray; Mitochondria; Oxydative stress; Rheumatoid arthritis.
Figures
References
-
- Nam JL, Ramiro S, Gaujoux-Viala C, Takase K, Leon-Garcia M, Emery P, et al. Efficacy of biological disease-modifying antirheumatic drugs: a systematic literature review informing the 2013 update of the EULAR recommendations for the management of rheumatoid arthritis. Ann Rheum Dis. 2014;73(3):516–28. doi: 10.1136/annrheumdis-2013-204577. - DOI - PubMed
-
- Orme ME, Macgilchrist KS, Mitchell S, Spurden D, Bird A. Systematic review and network meta-analysis of combination and monotherapy treatments in disease-modifying antirheumatic drug-experienced patients with rheumatoid arthritis: analysis of American College of Rheumatology criteria scores 20, 50, and 70. Biologics. 2012;6:429–64. - PMC - PubMed
-
- Ramiro S, Gaujoux-Viala C, Nam JL, Smolen JS, Buch M, Gossec L, et al. Safety of synthetic and biological DMARDs: a systematic literature review informing the 2013 update of the EULAR recommendations for management of rheumatoid arthritis. Ann Rheum Dis. 2014;73(3):529–35. doi: 10.1136/annrheumdis-2013-204575. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

