Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study
- PMID: 28545823
- PMCID: PMC5489698
- DOI: 10.1016/S1470-2045(17)30243-7
Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study
Abstract
Background: International consensus recognises four medulloblastoma molecular subgroups: WNT (MBWNT), SHH (MBSHH), group 3 (MBGrp3), and group 4 (MBGrp4), each defined by their characteristic genome-wide transcriptomic and DNA methylomic profiles. These subgroups have distinct clinicopathological and molecular features, and underpin current disease subclassification and initial subgroup-directed therapies that are underway in clinical trials. However, substantial biological heterogeneity and differences in survival are apparent within each subgroup, which remain to be resolved. We aimed to investigate whether additional molecular subgroups exist within childhood medulloblastoma and whether these could be used to improve disease subclassification and prognosis predictions.
Methods: In this retrospective cohort study, we assessed 428 primary medulloblastoma samples collected from UK Children's Cancer and Leukaemia Group (CCLG) treatment centres (UK), collaborating European institutions, and the UKCCSG-SIOP-PNET3 European clinical trial. An independent validation cohort (n=276) of archival tumour samples was also analysed. We analysed samples from patients with childhood medulloblastoma who were aged 0-16 years at diagnosis, and had central review of pathology and comprehensive clinical data. We did comprehensive molecular profiling, including DNA methylation microarray analysis, and did unsupervised class discovery of test and validation cohorts to identify consensus primary molecular subgroups and characterise their clinical and biological significance. We modelled survival of patients aged 3-16 years in patients (n=215) who had craniospinal irradiation and had been treated with a curative intent.
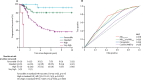
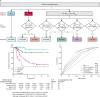
Findings: Seven robust and reproducible primary molecular subgroups of childhood medulloblastoma were identified. MBWNT remained unchanged and each remaining consensus subgroup was split in two. MBSHH was split into age-dependent subgroups corresponding to infant (<4·3 years; MBSHH-Infant; n=65) and childhood patients (≥4·3 years; MBSHH-Child; n=38). MBGrp3 and MBGrp4 were each split into high-risk (MBGrp3-HR [n=65] and MBGrp4-HR [n=85]) and low-risk (MBGrp3-LR [n=50] and MBGrp4-LR [n=73]) subgroups. These biological subgroups were validated in the independent cohort. We identified features of the seven subgroups that were predictive of outcome. Cross-validated subgroup-dependent survival models, incorporating these novel subgroups along with secondary clinicopathological and molecular features and established disease risk-factors, outperformed existing disease risk-stratification schemes. These subgroup-dependent models stratified patients into four clinical risk groups for 5-year progression-free survival: favourable risk (54 [25%] of 215 patients; 91% survival [95% CI 82-100]); standard risk (50 [23%] patients; 81% survival [70-94]); high-risk (82 [38%] patients; 42% survival [31-56]); and very high-risk (29 [13%] patients; 28% survival [14-56]).
Interpretation: The discovery of seven novel, clinically significant subgroups improves disease risk-stratification and could inform treatment decisions. These data provide a new foundation for future research and clinical investigations.
Funding: Cancer Research UK, The Tom Grahame Trust, Star for Harris, Action Medical Research, SPARKS, The JGW Patterson Foundation, The INSTINCT network (co-funded by The Brain Tumour Charity, Great Ormond Street Children's Charity, and Children with Cancer UK).
Copyright © 2017 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Figures




Comment in
-
Refining medulloblastoma subgroups.Lancet Oncol. 2017 Jul;18(7):847-848. doi: 10.1016/S1470-2045(17)30332-7. Epub 2017 May 22. Lancet Oncol. 2017. PMID: 28545822 No abstract available.
References
-
- Fattet S, Haberler C, Legoix P. Beta-catenin status in paediatric medulloblastomas: correlation of immunohistochemical expression with mutational status, genetic profiles, and clinical characteristics. J Pathol. 2009;218:86–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

