A tripeptidyl peptidase 1 is a binding partner of the Golgi pH regulator (GPHR) in Dictyostelium
- PMID: 28546289
- PMCID: PMC5536908
- DOI: 10.1242/dmm.029280
A tripeptidyl peptidase 1 is a binding partner of the Golgi pH regulator (GPHR) in Dictyostelium
Abstract
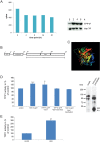
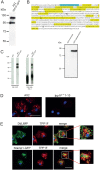
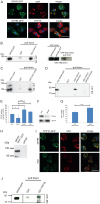
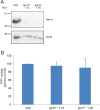
Mutations in tripeptidyl peptidase 1 (TPP1) have been associated with late infantile neuronal ceroid lipofuscinosis (NCL), a neurodegenerative disorder. TPP1 is a lysosomal serine protease, which removes tripeptides from the N-terminus of proteins and is composed of an N-terminal prodomain and a catalytic domain. It is conserved in mammals, amphibians, fish and the amoeba Dictyostelium discoideum. D. discoideum harbors at least six genes encoding TPP1, tpp1A to tpp1F We identified TPP1F as binding partner of Dictyostelium GPHR (Golgi pH regulator), which is an evolutionarily highly conserved intracellular transmembrane protein. A region encompassing the DUF3735 (GPHR_N) domain of GPHR was responsible for the interaction. In TPP1F, the binding site is located in the prodomain of the protein. The tpp1F gene is transcribed throughout development and translated into a polypeptide of ∼65 kDa. TPP1 activity was demonstrated for TPP1F-GFP immunoprecipitated from D. discoideum cells. Its activity could be inhibited by addition of the recombinant DUF3735 domain of GPHR. Knockout tpp1F mutants did not display any particular phenotype, and TPP1 activity was not abrogated, presumably because tpp1B compensates as it has the highest expression level of all the TPP1 genes during growth. The GPHR interaction was not restricted to TPP1F but occurred also with TPP1B. As previous reports show that the majority of the TPP1 mutations in NCL resulted in reduction or loss of enzyme activity, we suggest that Dicyostelium could be used as a model system in which to test new reagents that could affect the activity of the protein and ameliorate the disease.
Keywords: Endosomes; Golgi pH regulator (GPHR); Lysosomes; Neuronal ceroid lipofuscinosis (NCL); Tripeptidyl peptidase 1 (TPP1).
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

