An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize
- PMID: 28548655
- PMCID: PMC5685555
- DOI: 10.1038/nplants.2017.73
An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize
Abstract
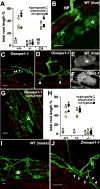
Most terrestrial plants, including crops, engage in beneficial interactions with arbuscular mycorrhizal fungi. Vital to the association is mutual recognition involving the release of diffusible signals into the rhizosphere. Previously, we identified the maize no perception 1 (nope1) mutant to be defective in early signalling. Here, we report cloning of ZmNope1 on the basis of synteny with rice. NOPE1 encodes a functional homologue of the Candida albicans N-acetylglucosamine (GlcNAc) transporter NGT1, and represents the first plasma membrane GlcNAc transporter identified from plants. In C. albicans, exposure to GlcNAc activates cell signalling and virulence. Similarly, in Rhizophagus irregularis treatment with rice wild-type but not nope1 root exudates induced transcriptome changes associated with signalling function, suggesting a requirement of NOPE1 function for presymbiotic fungal reprogramming.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Comment in
-
Arbuscular mycorrhiza: A new role for N-acetylglucosamine.Nat Plants. 2017 May 26;3:17085. doi: 10.1038/nplants.2017.85. Nat Plants. 2017. PMID: 28548654 No abstract available.
Similar articles
-
Transcriptional regulation of host NH₄⁺ transporters and GS/GOGAT pathway in arbuscular mycorrhizal rice roots.Plant Physiol Biochem. 2014 Feb;75:1-8. doi: 10.1016/j.plaphy.2013.11.029. Epub 2013 Dec 10. Plant Physiol Biochem. 2014. PMID: 24361504
-
A rice Serine/Threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane.Nat Commun. 2018 Nov 8;9(1):4677. doi: 10.1038/s41467-018-06865-z. Nat Commun. 2018. PMID: 30410018 Free PMC article.
-
A rice calcium-dependent protein kinase is expressed in cortical root cells during the presymbiotic phase of the arbuscular mycorrhizal symbiosis.BMC Plant Biol. 2011 May 19;11:90. doi: 10.1186/1471-2229-11-90. BMC Plant Biol. 2011. PMID: 21595879 Free PMC article.
-
Polyphony in the rhizosphere: presymbiotic communication in arbuscular mycorrhizal symbiosis.Curr Opin Plant Biol. 2013 Aug;16(4):473-9. doi: 10.1016/j.pbi.2013.06.005. Epub 2013 Jul 5. Curr Opin Plant Biol. 2013. PMID: 23834765 Review.
-
Conditioning plants for arbuscular mycorrhizal symbiosis through DWARF14-LIKE signalling.Curr Opin Plant Biol. 2021 Aug;62:102071. doi: 10.1016/j.pbi.2021.102071. Epub 2021 Jun 26. Curr Opin Plant Biol. 2021. PMID: 34186295 Free PMC article. Review.
Cited by
-
Phytohormones Regulate the Development of Arbuscular Mycorrhizal Symbiosis.Int J Mol Sci. 2018 Oct 12;19(10):3146. doi: 10.3390/ijms19103146. Int J Mol Sci. 2018. PMID: 30322086 Free PMC article. Review.
-
Plant Signaling and Metabolic Pathways Enabling Arbuscular Mycorrhizal Symbiosis.Plant Cell. 2017 Oct;29(10):2319-2335. doi: 10.1105/tpc.17.00555. Epub 2017 Aug 30. Plant Cell. 2017. PMID: 28855333 Free PMC article. Review.
-
A roadmap of plant membrane transporters in arbuscular mycorrhizal and legume-rhizobium symbioses.Plant Physiol. 2021 Dec 4;187(4):2071-2091. doi: 10.1093/plphys/kiab280. Plant Physiol. 2021. PMID: 34618047 Free PMC article. Review.
-
A glance at the 2017 edition of the international Molecular Mycorrhiza Meetings.Mycorrhiza. 2018 Feb;28(2):207-208. doi: 10.1007/s00572-017-0814-8. Mycorrhiza. 2018. PMID: 29274038 No abstract available.
-
Improvement of Verticillium Wilt Resistance by Applying Arbuscular Mycorrhizal Fungi to a Cotton Variety with High Symbiotic Efficiency under Field Conditions.Int J Mol Sci. 2018 Jan 13;19(1):241. doi: 10.3390/ijms19010241. Int J Mol Sci. 2018. PMID: 29342876 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

