Oligodendroglial myelination requires astrocyte-derived lipids
- PMID: 28549068
- PMCID: PMC5446120
- DOI: 10.1371/journal.pbio.1002605
Oligodendroglial myelination requires astrocyte-derived lipids
Abstract
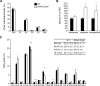
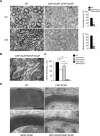
In the vertebrate nervous system, myelination of axons for rapid impulse propagation requires the synthesis of large amounts of lipids and proteins by oligodendrocytes and Schwann cells. Myelin membranes are thought to be cell-autonomously assembled by these axon-associated glial cells. Here, we report the surprising finding that in normal brain development, a substantial fraction of the lipids incorporated into central nervous system (CNS) myelin are contributed by astrocytes. The oligodendrocyte-specific inactivation of sterol regulatory element-binding protein (SREBP) cleavage-activating protein (SCAP), an essential coactivator of the transcription factor SREBP and thus of lipid biosynthesis, resulted in significantly retarded CNS myelination; however, myelin appeared normal at 3 months of age. Importantly, embryonic deletion of the same gene in astrocytes, or in astrocytes and oligodendrocytes, caused a persistent hypomyelination, as did deletion from astrocytes during postnatal development. Moreover, when astroglial lipid synthesis was inhibited, oligodendrocytes began incorporating circulating lipids into myelin membranes. Indeed, a lipid-enriched diet was sufficient to rescue hypomyelination in these conditional mouse mutants. We conclude that lipid synthesis by oligodendrocytes is heavily supplemented by astrocytes in vivo and that horizontal lipid flux is a major feature of normal brain development and myelination.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures




Similar articles
-
Ulk4 deficiency leads to hypomyelination in mice.Glia. 2018 Jan;66(1):175-190. doi: 10.1002/glia.23236. Epub 2017 Oct 16. Glia. 2018. PMID: 29034508
-
High-fat diet ameliorates neurological deficits caused by defective astrocyte lipid metabolism.FASEB J. 2012 Oct;26(10):4302-15. doi: 10.1096/fj.12-205807. Epub 2012 Jul 2. FASEB J. 2012. PMID: 22751013
-
Dysmyelination and reduced myelin basic protein gene expression by oligodendrocytes of SHP-1-deficient mice.J Neurosci Res. 2004 Jul 1;77(1):15-25. doi: 10.1002/jnr.20155. J Neurosci Res. 2004. PMID: 15197735
-
Iron Metabolism in Oligodendrocytes and Astrocytes, Implications for Myelination and Remyelination.ASN Neuro. 2020 Jan-Dec;12:1759091420962681. doi: 10.1177/1759091420962681. ASN Neuro. 2020. PMID: 32993319 Free PMC article. Review.
-
Severe Convulsions and Dysmyelination in Both Jimpy and Cx32/47 -/- Mice may Associate Astrocytic L-Channel Function with Myelination and Oligodendrocytic Connexins with Internodal Kv Channels.Neurochem Res. 2017 Jun;42(6):1747-1766. doi: 10.1007/s11064-017-2194-z. Epub 2017 Feb 18. Neurochem Res. 2017. PMID: 28214987 Review.
Cited by
-
Context-dependent regulation of Notch signaling in glial development and tumorigenesis.Sci Adv. 2023 Nov 10;9(45):eadi2167. doi: 10.1126/sciadv.adi2167. Epub 2023 Nov 10. Sci Adv. 2023. PMID: 37948517 Free PMC article.
-
Astrocytes and Inflammatory T Helper Cells: A Dangerous Liaison in Multiple Sclerosis.Front Immunol. 2022 Feb 8;13:824411. doi: 10.3389/fimmu.2022.824411. eCollection 2022. Front Immunol. 2022. PMID: 35211120 Free PMC article. Review.
-
Myelination and mTOR.Glia. 2018 Apr;66(4):693-707. doi: 10.1002/glia.23273. Epub 2017 Dec 6. Glia. 2018. PMID: 29210103 Free PMC article. Review.
-
LRP1 regulates peroxisome biogenesis and cholesterol homeostasis in oligodendrocytes and is required for proper CNS myelin development and repair.Elife. 2017 Dec 18;6:e30498. doi: 10.7554/eLife.30498. Elife. 2017. PMID: 29251594 Free PMC article.
-
Nicotinamide and Nicotinoyl-Gamma-Aminobutyric Acid as Neuroprotective Agents Against Type 1 Diabetes-Induced Nervous System Impairments in Rats.Neurochem Res. 2024 Nov 11;50(1):1. doi: 10.1007/s11064-024-04257-y. Neurochem Res. 2024. PMID: 39527359
References
-
- Barres BA (2008) The mystery and magic of glia: a perspective on their roles in health and disease. Neuron 60: 430–440. doi: 10.1016/j.neuron.2008.10.013 - DOI - PubMed
-
- Waxman SG (1980) Determinants of conduction velocity in myelinated nerve fibers. Muscle Nerve 3: 141–150. doi: 10.1002/mus.880030207 - DOI - PubMed
-
- Nave KA, Werner HB (2014) Myelination of the nervous system: mechanisms and functions. Annu Rev Cell Dev Biol 30: 503–533. doi: 10.1146/annurev-cellbio-100913-013101 - DOI - PubMed
-
- Norton WT, Poduslo SE (1973) Myelination in rat brain: changes in myelin composition during brain maturation. J Neurochem 21: 759–773. - PubMed
-
- Chrast R, Saher G, Nave KA, Verheijen MH (2011) Lipid metabolism in myelinating glial cells: lessons from human inherited disorders and mouse models. J Lipid Res 52: 419–434. doi: 10.1194/jlr.R009761 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

