Applications of Single-Molecule Methods to Membrane Protein Folding Studies
- PMID: 28549924
- PMCID: PMC5700846
- DOI: 10.1016/j.jmb.2017.05.021
Applications of Single-Molecule Methods to Membrane Protein Folding Studies
Abstract
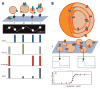
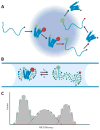
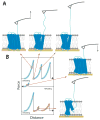
Protein folding is a fundamental life process with many implications throughout biology and medicine. Consequently, there have been enormous efforts to understand how proteins fold. Almost all of this effort has focused on water-soluble proteins, however, leaving membrane proteins largely wandering in the wilderness. The neglect has occurred not because membrane proteins are unimportant but rather because they present many theoretical and technical complications. Indeed, quantitative membrane protein folding studies are generally restricted to a handful of well-behaved proteins. Single-molecule methods may greatly alter this picture, however, because the ability to work at or near infinite dilution removes aggregation problems, one of the main technical challenges of membrane protein folding studies.
Keywords: atomic force spectroscopy; fluorescence; forced unfolding; magnetic tweezer.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

References
-
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

