Prevalence and Genomic Characterization of Escherichia coli O157:H7 in Cow-Calf Herds throughout California
- PMID: 28550057
- PMCID: PMC5541215
- DOI: 10.1128/AEM.00734-17
Prevalence and Genomic Characterization of Escherichia coli O157:H7 in Cow-Calf Herds throughout California
Abstract
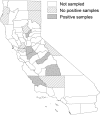
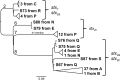
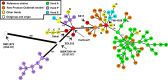
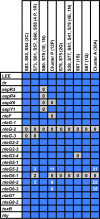
Escherichia coli serotype O157:H7 is a zoonotic food- and waterborne bacterial pathogen that causes a high hospitalization rate and can cause life-threatening complications. Increasingly, E. coli O157:H7 infections appear to originate from fresh produce. Ruminants, such as cattle, are a prominent reservoir of E. coli O157:H7 in the United States. California is one of the most agriculturally productive regions in the world for fresh produce, beef, and milk. The close proximity of fresh produce and cattle presents food safety challenges on a uniquely large scale. We performed a survey of E. coli O157:H7 on 20 farms in California to observe the regional diversity and prevalence of E. coli O157:H7. Isolates were obtained from enrichment cultures of cow feces. Some farms were sampled on two dates. Genomes from isolates were sequenced to determine their relatedness and pathogenic potential. E. coli O157:H7 was isolated from approximately half of the farms. The point prevalence of E. coli O157:H7 on farms was highly variable, ranging from zero to nearly 90%. Within farms, generally one or a few lineages were found, even when the rate of isolation was high. On farms with high isolation rates, a single clonal lineage accounted for most of the isolates. Farms that were visited months after the first visit might have had the same lineages of E. coli O157:H7. Strains of E. coli O157:H7 may be persistent for months on farms.IMPORTANCE This survey of 20 cow-calf operations from different regions of California provides an in depth look at resident Escherichia coli O157:H7 populations at the molecular level. E. coli O157:H7 is found to have a highly variable prevalence, and with whole-genome sequencing, high prevalences in herds were found to be due to a single lineage shed from multiple cows. Few repeat lineages were found between farms in this area; therefore, we predict that E. coli O157:H7 has significant diversity in this area beyond what is detected in this survey. All isolates from this study were found to have pathogenic potential based on the presence of key virulence gene sequences. This represents a novel insight into pathogen diversity within a single subtype and will inform future attempts to survey regional pathogen populations.
Keywords: California; Escherichia coli; O157:H7; STEC; cattle; food-borne pathogens; genomics.
Copyright © 2017 American Society for Microbiology.
Figures
Similar articles
-
Carriage and Subtypes of Foodborne Pathogens Identified in Wild Birds Residing near Agricultural Lands in California: a Repeated Cross-Sectional Study.Appl Environ Microbiol. 2020 Jan 21;86(3):e01678-19. doi: 10.1128/AEM.01678-19. Print 2020 Jan 21. Appl Environ Microbiol. 2020. PMID: 31757824 Free PMC article.
-
Results of a longitudinal study of the prevalence of Escherichia coli O157:H7 on cow-calf farms.Am J Vet Res. 2000 Nov;61(11):1375-9. doi: 10.2460/ajvr.2000.61.1375. Am J Vet Res. 2000. PMID: 11108182
-
Multiple-locus variable-nucleotide tandem repeat subtype analysis implicates European starlings as biological vectors for Escherichia coli O157:H7 in Ohio, USA.J Appl Microbiol. 2011 Oct;111(4):982-8. doi: 10.1111/j.1365-2672.2011.05102.x. Epub 2011 Aug 4. J Appl Microbiol. 2011. PMID: 21762472
-
Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products.J Anim Sci. 2007 Mar;85(13 Suppl):E63-72. doi: 10.2527/jas.2006-421. Epub 2006 Oct 23. J Anim Sci. 2007. PMID: 17060419 Review.
-
Perspectives on super-shedding of Escherichia coli O157:H7 by cattle.Foodborne Pathog Dis. 2015 Feb;12(2):89-103. doi: 10.1089/fpd.2014.1829. Epub 2014 Dec 16. Foodborne Pathog Dis. 2015. PMID: 25514549 Review.
Cited by
-
Persistent Circulation of Enterohemorrhagic Escherichia coli (EHEC) O157:H7 in Cattle Farms: Characterization of Enterohemorrhagic Escherichia coli O157:H7 Strains and Fecal Microbial Communities of Bovine Shedders and Non-shedders.Front Vet Sci. 2022 Mar 25;9:852475. doi: 10.3389/fvets.2022.852475. eCollection 2022. Front Vet Sci. 2022. PMID: 35411306 Free PMC article.
-
Longitudinal Study of Shiga Toxin-Producing Escherichia coli and Campylobacter jejuni on Finnish Dairy Farms and in Raw Milk.Appl Environ Microbiol. 2019 Mar 22;85(7):e02910-18. doi: 10.1128/AEM.02910-18. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30709824 Free PMC article.
-
Influence of Maternal BLV Infection on miRNA and tRF Expression in Calves.Pathogens. 2023 Nov 3;12(11):1312. doi: 10.3390/pathogens12111312. Pathogens. 2023. PMID: 38003777 Free PMC article.
-
Rates of evolutionary change of resident Escherichia coli O157:H7 differ within the same ecological niche.BMC Genomics. 2022 Apr 7;23(1):275. doi: 10.1186/s12864-022-08497-6. BMC Genomics. 2022. PMID: 35392797 Free PMC article.
-
Whole genome sequencing based typing and characterisation of Shiga-toxin producing Escherichia coli strains belonging to O157 and O26 serotypes and isolated in dairy farms.Ital J Food Saf. 2019 Feb 8;7(4):7673. doi: 10.4081/ijfs.2018.7673. eCollection 2018 Dec 31. Ital J Food Saf. 2019. PMID: 30854339 Free PMC article.
References
-
- United States Department of Agriculture. 2015. California agricultural statistics 2013 crop year. United States Department of Agriculture, National Agricultural Statistics Service, Pacific Regional Field Office, Sacramento, CA.
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

