A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer
- PMID: 28551381
- PMCID: PMC5478212
- DOI: 10.1016/j.ebiom.2017.04.022
A Specific Mutational Signature Associated with DNA 8-Oxoguanine Persistence in MUTYH-defective Colorectal Cancer
Abstract
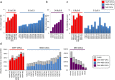
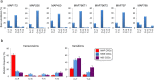
8-Oxoguanine, a common mutagenic DNA lesion, generates G:C>T:A transversions via mispairing with adenine during DNA replication. When operating normally, the MUTYH DNA glycosylase prevents 8-oxoguanine-related mutagenesis by excising the incorporated adenine. Biallelic MUTYH mutations impair this enzymatic function and are associated with colorectal cancer (CRC) in MUTYH-Associated Polyposis (MAP) syndrome. Here, we perform whole-exome sequencing that reveals a modest mutator phenotype in MAP CRCs compared to sporadic CRC stem cell lines or bulk tumours. The excess G:C>T:A transversion mutations in MAP CRCs exhibits a novel mutational signature, termed Signature 36, with a strong sequence dependence. The MUTYH mutational signature reflecting persistent 8-oxoG:A mismatches occurs frequently in the APC, KRAS, PIK3CA, FAT4, TP53, FAT1, AMER1, KDM6A, SMAD4 and SMAD2 genes that are associated with CRC. The occurrence of Signature 36 in other types of human cancer indicates that DNA 8-oxoguanine-related mutations might contribute to the development of cancer in other organs.
Keywords: 8-Oxoguanine; Base excision repair; Colorectal cancer; Exome sequencing; MUTYH-associated polyposis; Mutational signature.
Copyright © 2017 The Authors. Published by Elsevier B.V. All rights reserved.
Figures





References
-
- Alexandrov L.B. Understanding the origins of human cancer. Science. 2015;350:1175. - PubMed
-
- Al-Tassan N., Chmiel N.H., Maynard J., Fleming N., Livingston A.L., Williams G.T., Hodges A.K., Davies D.R., David S.S., Sampson J.R. Inherited variants of MYH associated with somatic G:C > T:A mutations in colorectal tumors. Nat. Genet. 2002;30:227–232. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

