Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence
- PMID: 28552492
- PMCID: PMC5462533
- DOI: 10.1016/j.cmet.2017.04.021
Age-Associated Loss of OPA1 in Muscle Impacts Muscle Mass, Metabolic Homeostasis, Systemic Inflammation, and Epithelial Senescence
Abstract
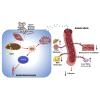
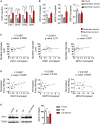
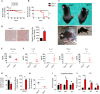
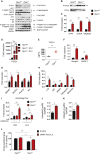
Mitochondrial dysfunction occurs during aging, but its impact on tissue senescence is unknown. Here, we find that sedentary but not active humans display an age-related decline in the mitochondrial protein, optic atrophy 1 (OPA1), that is associated with muscle loss. In adult mice, acute, muscle-specific deletion of Opa1 induces a precocious senescence phenotype and premature death. Conditional and inducible Opa1 deletion alters mitochondrial morphology and function but not DNA content. Mechanistically, the ablation of Opa1 leads to ER stress, which signals via the unfolded protein response (UPR) and FoxOs, inducing a catabolic program of muscle loss and systemic aging. Pharmacological inhibition of ER stress or muscle-specific deletion of FGF21 compensates for the loss of Opa1, restoring a normal metabolic state and preventing muscle atrophy and premature death. Thus, mitochondrial dysfunction in the muscle can trigger a cascade of signaling initiated at the ER that systemically affects general metabolism and aging.
Keywords: FGF21; FoxO; Opa1; aging; inflammation; mitochondria; muscle; oxidative stress; sarcopenia.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. - PubMed
-
- Amati-Bonneau P., Valentino M.L., Reynier P., Gallardo M.E., Bornstein B., Boissière A., Campos Y., Rivera H., de la Aleja J.G., Carroccia R. OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain. 2008;131:338–351. - PubMed
-
- Blaauw B., Canato M., Agatea L., Toniolo L., Mammucari C., Masiero E., Abraham R., Sandri M., Schiaffino S., Reggiani C. Inducible activation of Akt increases skeletal muscle mass and force without satellite cell activation. FASEB J. 2009;23:3896–3905. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

