The Recombinant Sea Urchin Immune Effector Protein, rSpTransformer-E1, Binds to Phosphatidic Acid and Deforms Membranes
- PMID: 28553283
- PMCID: PMC5427130
- DOI: 10.3389/fimmu.2017.00481
The Recombinant Sea Urchin Immune Effector Protein, rSpTransformer-E1, Binds to Phosphatidic Acid and Deforms Membranes
Abstract
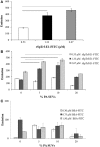
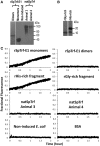
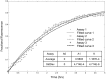
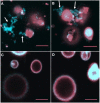
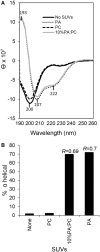
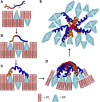
The purple sea urchin, Strongylocentrotus purpuratus, possesses a sophisticated innate immune system that functions without adaptive capabilities and responds to pathogens effectively by expressing the highly diverse SpTransformer gene family (formerly the Sp185/333 gene family). The swift gene expression response and the sequence diversity of SpTransformer cDNAs suggest that the encoded proteins have immune functions. Individual sea urchins can express up to 260 distinct SpTransformer proteins, and their diversity suggests that different versions may have different functions. Although the deduced proteins are diverse, they share an overall structure of a hydrophobic leader, a glycine-rich N-terminal region, a histidine-rich region, and a C-terminal region. Circular dichroism analysis of a recombinant SpTransformer protein, rSpTransformer-E1 (rSpTrf-E1) demonstrates that it is intrinsically disordered and transforms to α helical in the presence of buffer additives and binding targets. Although native SpTrf proteins are associated with the membranes of perinuclear vesicles in the phagocyte class of coelomocytes and are present on the surface of small phagocytes, they have no predicted transmembrane region or conserved site for glycophosphatidylinositol linkage. To determine whether native SpTrf proteins associate with phagocyte membranes through interactions with lipids, when rSpTrf-E1 is incubated with lipid-embedded nylon strips, it binds to phosphatidic acid (PA) through both the glycine-rich region and the histidine-rich region. Synthetic liposomes composed of PA and phosphatidylcholine show binding between rSpTrf-E1 and PA by fluorescence resonance energy transfer, which is associated with leakage of luminal contents suggesting changes in lipid organization and perhaps liposome lysis. Interactions with liposomes also change membrane curvature leading to liposome budding, fusion, and invagination, which is associated with PA clustering induced by rSpTrf-E1 binding. Longer incubations result in the extraction of PA from the liposomes, which form disorganized clusters. CD shows that when rSpTrf-E1 binds to PA, it changes its secondary structure from disordered to α helical. These results provide evidence for how SpTransformer proteins may associate with molecules that have exposed phosphates including PA on cell membranes and how the characteristic of protein multimerization may drive changes in the organization of membrane lipids.
Keywords: Sp185/333; conformational plasticity; echinoderm; innate immunity; lipid clusters; liposomes.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

