Designing logical codon reassignment - Expanding the chemistry in biology
- PMID: 28553457
- PMCID: PMC5424465
- DOI: 10.1039/c4sc01534g
Designing logical codon reassignment - Expanding the chemistry in biology
Abstract
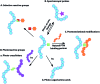
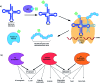
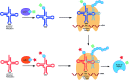
Over the last decade, the ability to genetically encode unnatural amino acids (UAAs) has evolved rapidly. The programmed incorporation of UAAs into recombinant proteins relies on the reassignment or suppression of canonical codons with an amino-acyl tRNA synthetase/tRNA (aaRS/tRNA) pair, selective for the UAA of choice. In order to achieve selective incorporation, the aaRS should be selective for the designed tRNA and UAA over the endogenous amino acids and tRNAs. Enhanced selectivity has been achieved by transferring an aaRS/tRNA pair from another kingdom to the organism of interest, and subsequent aaRS evolution to acquire enhanced selectivity for the desired UAA. Today, over 150 non-canonical amino acids have been incorporated using such methods. This enables the introduction of a large variety of structures into proteins, in organisms ranging from prokaryote, yeast and mammalian cells lines to whole animals, enabling the study of protein function at a level that could not previously be achieved. While most research to date has focused on the suppression of 'non-sense' codons, recent developments are beginning to open up the possibility of quadruplet codon decoding and the more selective reassignment of sense codons, offering a potentially powerful tool for incorporating multiple amino acids. Here, we aim to provide a focused review of methods for UAA incorporation with an emphasis in particular on the different tRNA synthetase/tRNA pairs exploited or developed, focusing upon the different UAA structures that have been incorporated and the logic behind the design and future creation of such systems. Our hope is that this will help rationalize the design of systems for incorporation of unexplored unnatural amino acids, as well as novel applications for those already known.
Figures

References
-
- Crick F. Nature. 1970;227:561. - PubMed
-
- Lodish H., Berk A., Zipursky S. L., Matsudaira P., Baltimore D. and Darnell J., Molecular Cell Biology, W. H. Freeman, New York, 2000.
-
- Martin A. B., Schultz P. G. Trends Cell Biol. 1999;9:M24. - PubMed
-
- Liu C. C., Schultz P. G. Annu. Rev. Biochem. 2010;79:413. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

