Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation
- PMID: 28553941
- PMCID: PMC5658125
- DOI: 10.1038/nbt.3879
Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation
Abstract
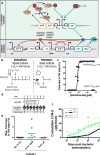
Bacteria can be engineered to function as diagnostics or therapeutics in the mammalian gut but commercial translation of technologies to accomplish this has been hindered by the susceptibility of synthetic genetic circuits to mutation and unpredictable function during extended gut colonization. Here, we report stable, engineered bacterial strains that maintain their function for 6 months in the mouse gut. We engineered a commensal murine Escherichia coli strain to detect tetrathionate, which is produced during inflammation. Using our engineered diagnostic strain, which retains memory of exposure in the gut for analysis by fecal testing, we detected tetrathionate in both infection-induced and genetic mouse models of inflammation over 6 months. The synthetic genetic circuits in the engineered strain were genetically stable and functioned as intended over time. The durable performance of these strains confirms the potential of engineered bacteria as living diagnostics.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
-
- Clairmont C, et al. Biodistribution and genetic stability of the novel antitumor agent VNP20009, a genetically modified strain of Salmonella typhimurium. J Infect Dis. 2000;181:1996–2002. - PubMed
-
- Ceroni F, Algar R, Stan GB, Ellis T. Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods. 2015;12:415–418. - PubMed
-
- Sleight SC, Sauro HM. Visualization of Evolutionary Stability Dynamics and Competitive Fitness of Escherichia coli Engineered with Randomized Multigene Circuits. ACS Synth Biol. 2013;2:519–528. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

