Structural basis for high-affinity fluorophore binding and activation by RNA Mango
- PMID: 28553947
- PMCID: PMC5550021
- DOI: 10.1038/nchembio.2392
Structural basis for high-affinity fluorophore binding and activation by RNA Mango
Abstract
Genetically encoded fluorescent protein tags have revolutionized proteome studies, whereas the lack of intrinsically fluorescent RNAs has hindered transcriptome exploration. Among several RNA-fluorophore complexes that potentially address this problem, RNA Mango has an exceptionally high affinity for its thiazole orange (TO)-derived fluorophore, TO1-Biotin (Kd ∼3 nM), and, in complex with related ligands, it is one of the most redshifted fluorescent macromolecular tags known. To elucidate how this small aptamer exhibits such properties, which make it well suited for studying low-copy cellular RNAs, we determined its 1.7-Å-resolution co-crystal structure. Unexpectedly, the entire ligand, including TO, biotin and the linker connecting them, abuts one of the near-planar faces of the three-tiered G-quadruplex. The two heterocycles of TO are held in place by two loop adenines and form a 45° angle with respect to each other. Minimizing this angle would increase quantum yield and further improve this tool for in vivo RNA visualization.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
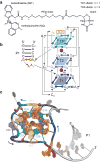
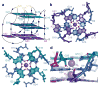


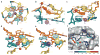
References
-
- Dolgosheina EV, et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol. 2014;9:2412–2420. - PubMed
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. - PubMed
-
- Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125:14716–14717. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

