ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner
- PMID: 28553953
- PMCID: PMC5633655
- DOI: 10.1038/onc.2017.146
ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner
Abstract
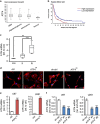
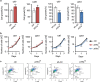
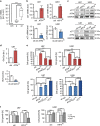
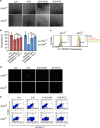
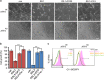
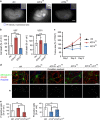
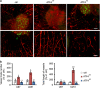
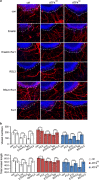
Activating transcription factor 4 (ATF4) is a critical mediator of metabolic and oxidative homeostasis and cell survival. ATF4 is elevated in response to diverse microenvironmental stresses, including starvation, ER stress damages and exposure to toxic factors. Here we show that ATF4 expression fosters the malignancy of primary brain tumors (WHO grade III and IV gliomas) and increases proliferation and tumor angiogenesis. Hence, ATF4 expression promotes cell migration and anchorage-independent cell growth, whereas siRNA-mediated knockdown of ATF4 attenuates these features of malignancy in human gliomas. Further experiments revealed that ATF4-dependent tumor promoting effects are mediated by transcriptional targeting the glutamate antiporter xCT/SCL7A11 (also known as system Xc-). Thus, xCT is elevated as a consequence of ATF4 activation. We further found evidence that ATF4-induced proliferation can be attenuated by pharmacological or genetic xCT inhibition and ferroptosis inducers such as sorafenib, erastin and GPx4 inhibitor RSL3. Further, fostered xCT expression promotes cell survival and growth in ATF4 knockdown cells. Moreover, increased xCT levels ameliorate sorafenib and erastin-induced ferroptosis. Conversely, ATF4 knockdown renders cells susceptible for erastin, sorafenib and RSL3-induced ferroptosis. We further identified that ATF4 promotes tumor-mediated neuronal cell death which can be alleviated by xCT inhibition. Moreover, elevated ATF4 expression in gliomas promotes tumor angiogenesis. Noteworthy, ATF4-induced angiogenesis could be diminished by ferroptosis inducers erastin and by GPx4 inhibitor RSL3. Our data provide proof-of-principle evidence that ATF4 fosters proliferation and induces a toxic microenvironmental niche. Furthermore, ATF4 increases tumor angiogenesis and shapes the vascular architecture in a xCT-dependent manner. Thus, inhibition of ATF4 is a valid target for diminishing tumor growth and vasculature via sensitizing tumor cells for ferroptosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Eyupoglu IY, Buchfelder M, Savaskan NE. Surgical resection of malignant gliomas-role in optimizing patient outcome. Nat Rev Neurol 2013; 9: 141–151. - PubMed
-
- Chevet E, Hetz C, Samali A. Endoplasmic reticulum stress-activated cell reprogramming in oncogenesis. Cancer Discov 2015; 5: 586–597. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

