ER-plasma membrane junctions: Why and how do we study them?
- PMID: 28554772
- PMCID: PMC5542405
- DOI: 10.1016/j.bbamcr.2017.05.018
ER-plasma membrane junctions: Why and how do we study them?
Abstract
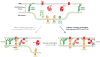
Endoplasmic reticulum (ER)-plasma membrane (PM) junctions are membrane microdomains important for communication between the ER and the PM. ER-PM junctions were first reported in muscle cells in 1957, but mostly ignored in non-excitable cells due to their scarcity and lack of functional significance. In 2005, the discovery of stromal interaction molecule 1 (STIM1) mediating a universal Ca2+ feedback mechanism at ER-PM junctions in mammalian cells led to a resurgence of research interests toward ER-PM junctions. In the past decade, several major advancements have been made in this emerging topic in cell biology, including the generation of tools for labeling ER-PM junctions and the unraveling of mechanisms underlying regulation and functions of ER-PM junctions. This review summarizes early studies, recently developed tools, and current advances in the characterization and understanding of ER-PM junctions. This article is part of a Special Issue entitled: Membrane Contact Sites edited by Christian Ungermann and Benoit Kornmann.
Copyright © 2017 Elsevier B.V. All rights reserved.
Figures




References
-
- Baumann O, Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. International review of cytology. 2001;205:149–214. - PubMed
-
- Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

