The serine/threonine phosphatases of apicomplexan parasites
- PMID: 28556455
- PMCID: PMC5787372
- DOI: 10.1111/mmi.13715
The serine/threonine phosphatases of apicomplexan parasites
Abstract
The balance between phosphorylation and de-phosphorylation, which is delicately regulated by protein kinases and phosphatases, is critical for nearly all biological processes. The Apicomplexa are a large phylum which contains various parasitic protists, including human pathogens, such as Plasmodium, Toxoplasma, Cryptosporidium and Babesia species. The diverse life cycles of these parasites are highly complex and, not surprisingly, many of their key steps are exquisitely regulated by phosphorylation. Interestingly, many of the kinases and phosphatases, as well as the substrates involved in these events are unique to the parasites and therefore phosphorylation constitutes a viable target for antiparasitic intervention. Most progress on this realm has come from studies in Toxoplasma and Plasmodium of their respective kinomes and phosphoproteomes. Nonetheless, given their likely importance, phosphatases have recently become the focus of research within the apicomplexan parasites. In this review, we concentrate on serine/threonine phosphatases in apicomplexan parasites, with the focus on comprehensively identifying and naming protein phosphatases in available apicomplexan genomes, and summarizing the progress of their functional analyses in recent years.
© 2017 John Wiley & Sons Ltd.
Figures


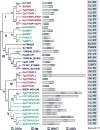
References
-
- Alvarez CA, Suvorova ES. A checkpoint roadmap for the complex cell division of Apicomplexa parasites. bioRxiv 2017
-
- Azevedo MF, Sanders PR, Krejany E, Nie CQ, Fu P, Bach LA, Wunderlich G, Crabb BS, Gilson PR. Inhibition of Plasmodium falciparum CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J. 2013;452:433–441. doi: 410.1042/BJ20130124. - PubMed
-
- Bansal A, Singh S, More KR, Hans D, Nangalia K, Yogavel M, Sharma A, Chitnis CE. Characterization of Plasmodium falciparum calcium-dependent protein kinase 1 (PfCDPK1) and its role in microneme secretion during erythrocyte invasion. J Biol Chem. 2013;288:1590–1602. doi: 1510.1074/jbc.M1112.411934. Epub 412012 Nov 411930. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

