Comparative molecular profiling of HPV-induced squamous cell carcinomas
- PMID: 28556593
- PMCID: PMC5504316
- DOI: 10.1002/cam4.1108
Comparative molecular profiling of HPV-induced squamous cell carcinomas
Abstract
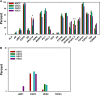
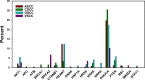
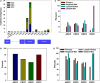
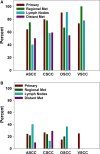
Approximately 5% of all cancer incidences result from human papillomavirus (HPV) infection. HPV infection most commonly leads to cancers of the anogenital region or oropharynx. It is unknown whether different HPV-mediated cancers collectively share a molecular signature and it is important to determine if there are targetable alterations common to different types of HPV-positive tumors. We analyzed 743 p53 wild-type samples of anal, cervical, oropharyngeal, and vulvar squamous cell carcinomas which underwent multiplatform testing at a commercial molecular profiling service. Expression of 24 proteins was measured by immunohistochemistry (IHC), mutation of 48 genes was determined by next-generation and Sanger sequencing, and copy number alteration for six genes was determined by in situ hybridization. The four cohorts had remarkably similar molecular profiles. No gene had a statistically significant difference in mutation frequency or copy number change between the four different types of squamous cell carcinomas. The only significant differences between cohorts were frequency of ERCC1 and SPARC loss as determined by IHC. In all four cancer types, oncogene mutation and PD-L1 expression was relatively infrequent. The most commonly mutated gene was PIK3CA, with mutations most often affecting the helical domain of the protein and accompanied by concurrent lack of PTEN expression. Loss of MGMT and RRM1 was common among the four cohorts and may be predictive of response to cytotoxic therapies not currently being used to treat these cancer types. The similar molecular profiles of the four cohorts indicate that treatment strategies may be similarly efficacious across HPV-positive cancers.
Keywords: HPV; Biomarkers; DNA sequencing; molecular profiling; protein expression; squamous cell carcinoma.
© 2017 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures
Similar articles
-
Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing.Ann Oncol. 2015 Jun;26(6):1216-1223. doi: 10.1093/annonc/mdv109. Epub 2015 Feb 23. Ann Oncol. 2015. PMID: 25712460 Free PMC article.
-
Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas.Clin Cancer Res. 2015 Feb 1;21(3):632-41. doi: 10.1158/1078-0432.CCR-13-3310. Epub 2014 Jul 23. Clin Cancer Res. 2015. PMID: 25056374 Free PMC article.
-
Molecular subclassification determined by human papillomavirus and epidermal growth factor receptor status is associated with the prognosis of oropharyngeal squamous cell carcinoma.Hum Pathol. 2016 Apr;50:51-61. doi: 10.1016/j.humpath.2015.11.001. Epub 2015 Nov 17. Hum Pathol. 2016. PMID: 26997438
-
HPV-related squamous cell carcinoma of the head and neck: An update on testing in routine pathology practice.Semin Diagn Pathol. 2015 Sep;32(5):344-51. doi: 10.1053/j.semdp.2015.02.013. Epub 2015 Feb 4. Semin Diagn Pathol. 2015. PMID: 25724476 Review.
-
Pathogenesis of Penile Squamous Cell Carcinoma: Molecular Update and Systematic Review.Int J Mol Sci. 2021 Dec 27;23(1):251. doi: 10.3390/ijms23010251. Int J Mol Sci. 2021. PMID: 35008677 Free PMC article.
Cited by
-
Comparison of clinicopathological and genomic profiles in anal squamous cell carcinoma between Japanese and Caucasian cohorts.Sci Rep. 2023 Mar 3;13(1):3587. doi: 10.1038/s41598-023-30624-w. Sci Rep. 2023. PMID: 36869079 Free PMC article.
-
RNA-seq analysis identifies transcriptomic profiles associated with anal cancer recurrence among people living with HIV.Ann Med. 2023 Dec;55(1):2199366. doi: 10.1080/07853890.2023.2199366. Ann Med. 2023. PMID: 37177979 Free PMC article.
-
Pathological variants in HPV-independent vulvar tumours.Sci Rep. 2025 Jan 9;15(1):1486. doi: 10.1038/s41598-024-84688-3. Sci Rep. 2025. PMID: 39789097 Free PMC article.
-
Lack of Conserved miRNA Deregulation in HPV-Induced Squamous Cell Carcinomas.Biomolecules. 2021 May 20;11(5):764. doi: 10.3390/biom11050764. Biomolecules. 2021. PMID: 34065237 Free PMC article.
-
Prevalence and prognostic value of PD-L1 expression and tumor mutational burden in persistent, recurrent, or metastatic cervical cancer.J Gynecol Oncol. 2024 Nov;35(6):e105. doi: 10.3802/jgo.2024.35.e105. Epub 2024 May 23. J Gynecol Oncol. 2024. PMID: 38857910 Free PMC article.
References
-
- Galloway, D. A. , Gewin L. C., Myers H., Luo W., Grandori C., Katzenellenbogen R. A., et al. 2005. Regulation of telomerase by human papillomaviruses. Cold Spring Harb. Symp. Quant. Biol. 70:209–215. - PubMed
-
- Felsani, A. , Mileo A. M., and Paggi M. G.. 2006. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 25:5277–5285. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

