Numerical integration methods and layout improvements in the context of dynamic RNA visualization
- PMID: 28558664
- PMCID: PMC5450055
- DOI: 10.1186/s12859-017-1682-0
Numerical integration methods and layout improvements in the context of dynamic RNA visualization
Abstract
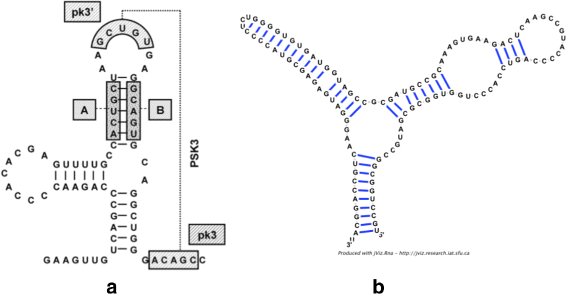
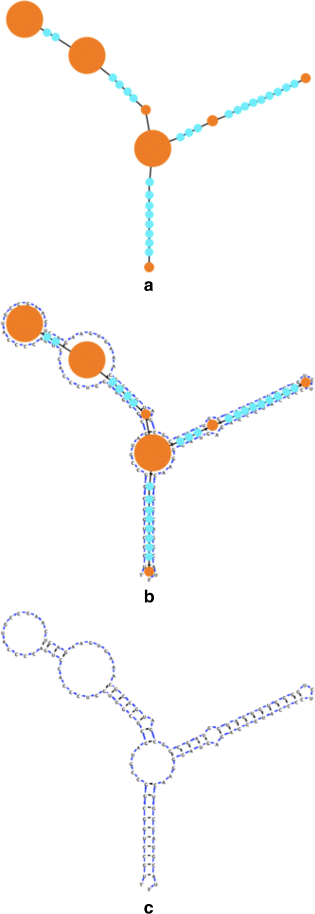
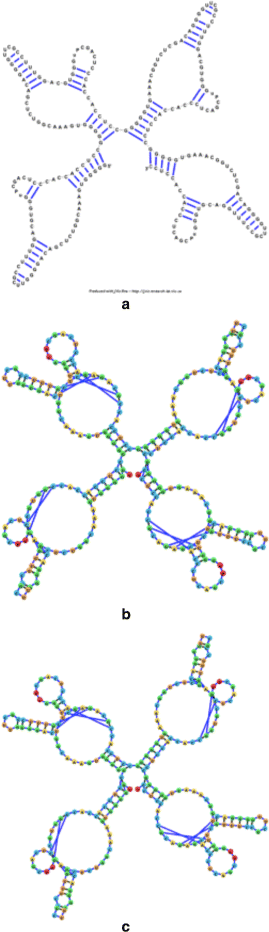
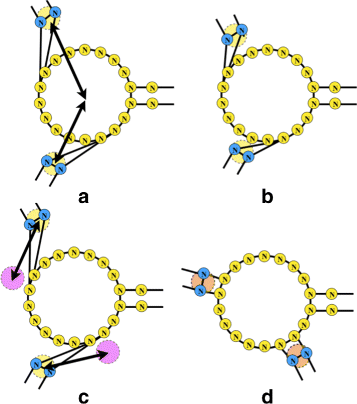
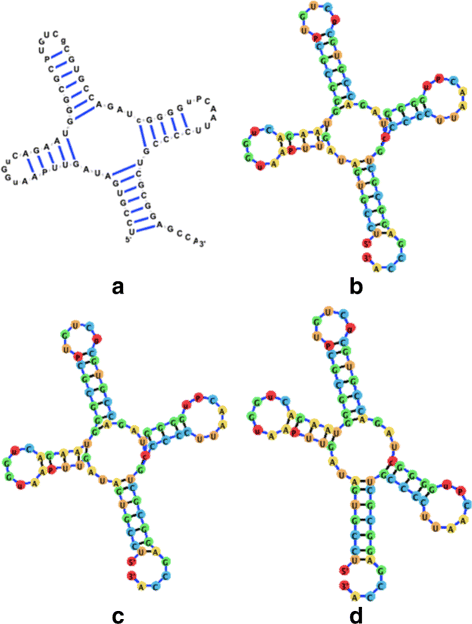
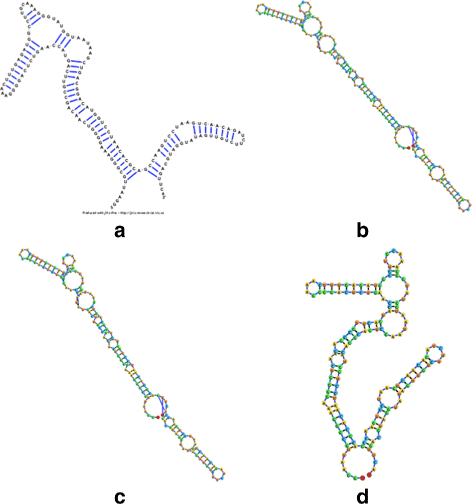
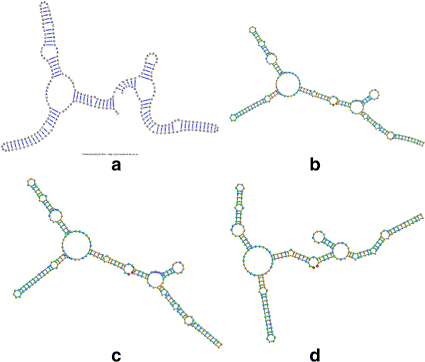
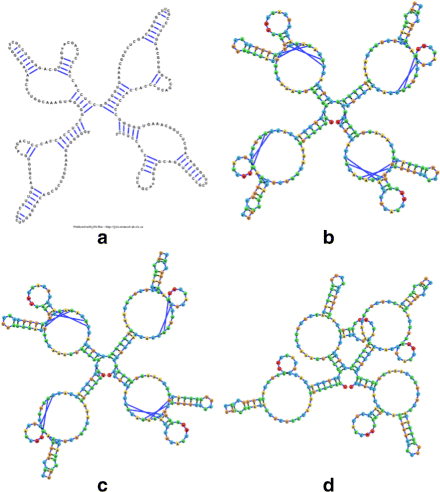
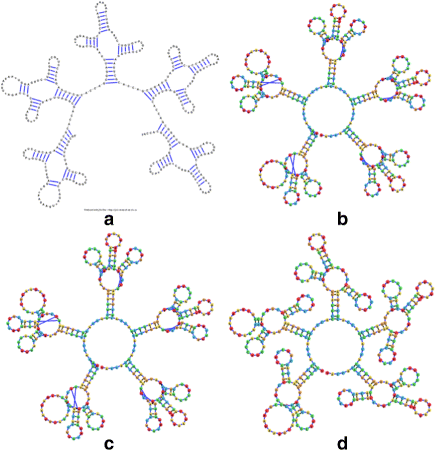
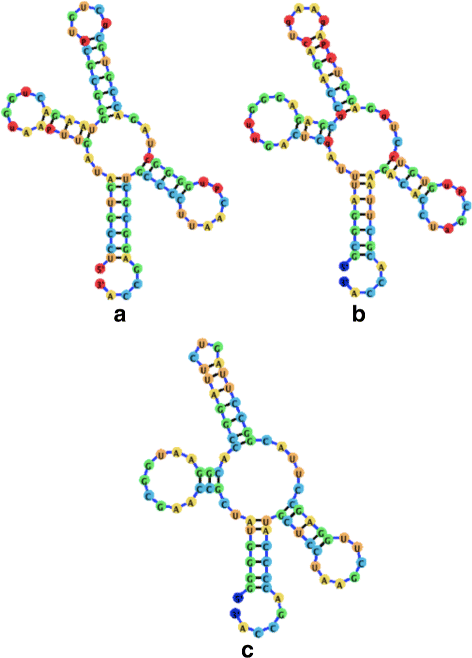
Background: RNA visualization software tools have traditionally presented a static visualization of RNA molecules with limited ability for users to interact with the resulting image once it is complete. Only a few tools allowed for dynamic structures. One such tool is jViz.RNA. Currently, jViz.RNA employs a unique method for the creation of the RNA molecule layout by mapping the RNA nucleotides into vertexes in a graph, which we call the detailed graph, and then utilizes a Newtonian mechanics inspired system of forces to calculate a layout for the RNA molecule. The work presented here focuses on improvements to jViz.RNA that allow the drawing of RNA secondary structures according to common drawing conventions, as well as dramatic run-time performance improvements. This is done first by presenting an alternative method for mapping the RNA molecule into a graph, which we call the compressed graph, and then employing advanced numerical integration methods for the compressed graph representation.
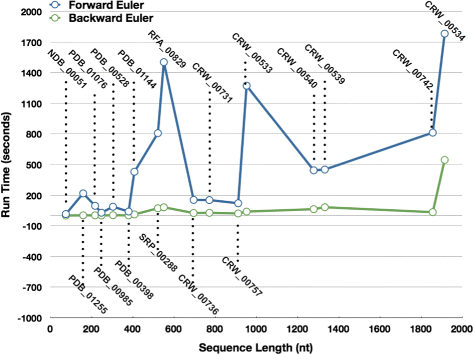
Results: Comparing the compressed graph and detailed graph implementations, we find that the compressed graph produces results more consistent with RNA drawing conventions. However, we also find that employing the compressed graph method requires a more sophisticated initial layout to produce visualizations that would require minimal user interference. Comparing the two numerical integration methods demonstrates the higher stability of the Backward Euler method, and its resulting ability to handle much larger time steps, a high priority feature for any software which entails user interaction.
Conclusion: The work in this manuscript presents the preferred use of compressed graphs to detailed ones, as well as the advantages of employing the Backward Euler method over the Forward Euler method. These improvements produce more stable as well as visually aesthetic representations of the RNA secondary structures. The results presented demonstrate that both the compressed graph representation, as well as the Backward Euler integrator, greatly enhance the run-time performance and usability. The newest iteration of jViz.RNA is available at https://jviz.cs.sfu.ca/download/download.html .
Keywords: Graph layout; Numerical integration; RNA; Visualization.
Figures



Similar articles
-
jViz.RNA 4.0-Visualizing pseudoknots and RNA editing employing compressed tree graphs.PLoS One. 2019 May 6;14(5):e0210281. doi: 10.1371/journal.pone.0210281. eCollection 2019. PLoS One. 2019. PMID: 31059508 Free PMC article.
-
JViz.Rna--a Java tool for RNA secondary structure visualization.IEEE Trans Nanobioscience. 2005 Sep;4(3):212-8. doi: 10.1109/tnb.2005.853646. IEEE Trans Nanobioscience. 2005. PMID: 16220684
-
TRAVeLer: a tool for template-based RNA secondary structure visualization.BMC Bioinformatics. 2017 Nov 15;18(1):487. doi: 10.1186/s12859-017-1885-4. BMC Bioinformatics. 2017. PMID: 29141608 Free PMC article.
-
Graph Visualization: Alternative Models Inspired by Bioinformatics.Sensors (Basel). 2023 Apr 4;23(7):3747. doi: 10.3390/s23073747. Sensors (Basel). 2023. PMID: 37050807 Free PMC article. Review.
-
Graph Theoretical Methods and Workflows for Searching and Annotation of RNA Tertiary Base Motifs and Substructures.Int J Mol Sci. 2021 Aug 9;22(16):8553. doi: 10.3390/ijms22168553. Int J Mol Sci. 2021. PMID: 34445259 Free PMC article. Review.
Cited by
-
jViz.RNA 4.0-Visualizing pseudoknots and RNA editing employing compressed tree graphs.PLoS One. 2019 May 6;14(5):e0210281. doi: 10.1371/journal.pone.0210281. eCollection 2019. PLoS One. 2019. PMID: 31059508 Free PMC article.
-
RiboSketch: versatile visualization of multi-stranded RNA and DNA secondary structure.Bioinformatics. 2018 Dec 15;34(24):4297-4299. doi: 10.1093/bioinformatics/bty468. Bioinformatics. 2018. PMID: 29912310 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

