Structure-based identification of inhibitors targeting obstruction of the HIVgp41 N-heptad repeat trimer
- PMID: 28558972
- PMCID: PMC5551449
- DOI: 10.1016/j.bmcl.2017.05.020
Structure-based identification of inhibitors targeting obstruction of the HIVgp41 N-heptad repeat trimer
Abstract
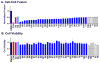
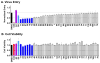
The viral protein HIVgp41 is an attractive and validated drug target that proceeds through a sequence of conformational changes crucial for membrane fusion, which facilitates viral entry. Prior work has identified inhibitors that interfere with the formation of a required six-helix bundle, composed of trimeric C-heptad (CHR) and N-heptad (NHR) repeat elements, through blocking association of an outer CHR helix or obstructing formation of the inner NHR trimer itself. In this work, we employed similarity-based scoring to identify and experimentally characterize 113 compounds, related to 2 small-molecule inhibitors recently reported by Allen et al. (Bioorg. Med. Chem Lett.2015, 25 2853-59), proposed to act via the NHR trimer obstruction mechanism. The compounds were first tested in an HIV cell-cell fusion assay with the most promising evaluated in a second, more biologically relevant viral entry assay. Of the candidates, compound #11 emerged as the most promising hit (IC50=37.81µM), as a result of exhibiting activity in both assays with low cytotoxicity, as was similarly seen with the known control peptide inhibitor C34. The compound also showed no inhibition of VSV-G pseudotyped HIV entry compared to a control inhibitor suggesting it was specific for HIVgp41. Molecular dynamics simulations showed the predicted DOCK pose of #11 interacts with HIVgp41 in an energetic fashion (per-residue footprints) similar to the four native NHR residues (IQLT) which candidate inhibitors were intended to mimic.
Keywords: Computer-aided drug design; DOCK; Docking; Footprint similarity; HIV; Hungarian similarity; Structure-based drug design; Viral entry; Virtual screening; gp41.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures
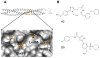


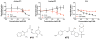

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

