Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease
- PMID: 28559085
- PMCID: PMC5565704
- DOI: 10.1016/j.ophtha.2017.04.008
Clinically Focused Molecular Investigation of 1000 Consecutive Families with Inherited Retinal Disease
Abstract
Purpose: To devise a comprehensive multiplatform genetic testing strategy for inherited retinal disease and to describe its performance in 1000 consecutive families seen by a single clinician.
Design: Retrospective series.
Participants: One thousand consecutive families seen by a single clinician.
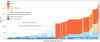
Methods: The clinical records of all patients seen by a single retina specialist between January 2010 and June 2016 were reviewed, and all patients who met the clinical criteria for a diagnosis of inherited retinal disease were included in the study. Each patient was assigned to 1 of 62 diagnostic categories, and this clinical diagnosis was used to define the scope and order of the molecular investigations that were performed. The number of nucleotides evaluated in a given subject ranged from 2 to nearly 900 000.
Main outcome measures: Sensitivity and false genotype rate.
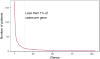
Results: Disease-causing genotypes were identified in 760 families (76%). These genotypes were distributed across 104 different genes. More than 75% of these 104 genes have coding sequences small enough to be packaged efficiently into an adeno-associated virus. Mutations in ABCA4 were the most common cause of disease in this cohort (173 families), whereas mutations in 80 genes caused disease in 5 or fewer families (i.e., 0.5% or less). Disease-causing genotypes were identified in 576 of the families without next-generation sequencing (NGS). This included 23 families with mutations in the repetitive region of RPGR exon 15 that would have been missed by NGS. Whole-exome sequencing of the remaining 424 families revealed mutations in an additional 182 families, and whole-genome sequencing of 4 of the remaining 242 families revealed 2 additional genotypes that were invisible by the other methods. Performing the testing in a clinically focused tiered fashion would be 6.1% more sensitive and 17.7% less expensive and would have a significantly lower average false genotype rate than using whole-exome sequencing to assess more than 300 genes in all patients (7.1% vs. 128%; P < 0.001).
Conclusions: Genetic testing for inherited retinal disease is now more than 75% sensitive. A clinically directed tiered testing strategy can increase sensitivity and improve statistical significance without increasing cost.
Copyright © 2017 American Academy of Ophthalmology. Published by Elsevier Inc. All rights reserved.
Figures





Comment in
-
Genetic Testing for Inherited Retinal Disease.Ophthalmology. 2017 Sep;124(9):1254-1255. doi: 10.1016/j.ophtha.2017.06.018. Ophthalmology. 2017. PMID: 28823343 No abstract available.
-
Inherited Retinal Disease Panels-Caveat Emptor-Truly Know Your Inherited Retinal Disease Panel.Retina. 2022 Jan 1;42(1):1-3. doi: 10.1097/IAE.0000000000003319. Retina. 2022. PMID: 34690342 Free PMC article. No abstract available.
References
-
- Cavenee WK, Dryja TP, Phillips RA, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. - PubMed
-
- Friend SH, Bernards R, Rogelj S, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. - PubMed
-
- Dryja TP, McGee TL, Reichel E, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

